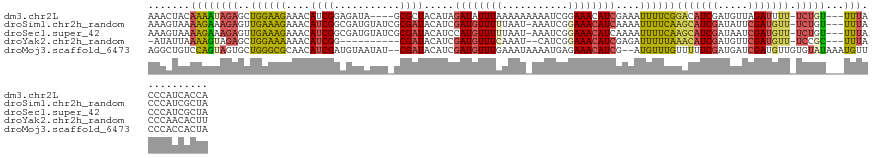
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 22,471,201 – 22,471,323 |
| Length | 122 |
| Max. P | 0.996860 |

| Location | 22,471,201 – 22,471,323 |
|---|---|
| Length | 122 |
| Sequences | 5 |
| Columns | 130 |
| Reading direction | forward |
| Mean pairwise identity | 71.72 |
| Shannon entropy | 0.49452 |
| G+C content | 0.35196 |
| Mean single sequence MFE | -31.94 |
| Consensus MFE | -12.69 |
| Energy contribution | -12.97 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.992900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
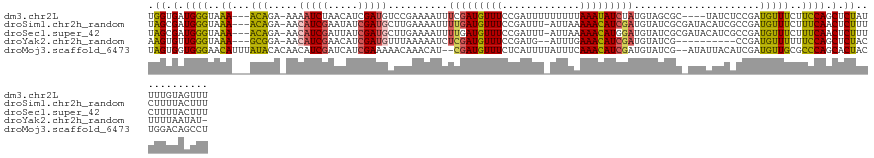
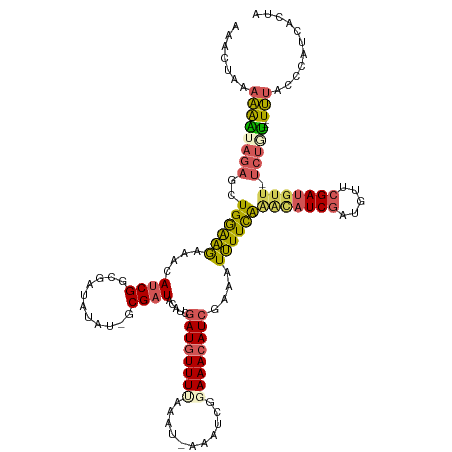
>dm3.chr2L 22471201 122 + 23011544 UGGUGAUGGGUAAA---ACAGA-AAAAUCUAACAUCGAUGUCCGAAAAUUUCGAUGUUUCCGAUUUUUUUUUAAAUAUCUAUGUAGCGC----UAUCUCCGAUGUUUCUUCCAGCUCUAUUUUGUAGUUU .....(((((((..---..(((-((((((.((((((((.((......)).))))))))...))))))))).....)))))))((((.((----(......((....))....))).)))).......... ( -22.80, z-score = -0.64, R) >droSim1.chr2h_random 3518 125 - 3178526 UAGCGAUGGGUAAA---ACAGA-AACAUCGAAUAUCGAUGCUUGAAAAUUUUGAUGUUUCCGAUUU-AUUAAAAACAUCGAUGUAUCGCGAUACAUCGCCGAUGUUUCUUUCAACUCUUUCUUUUACUUU ....((.((((..(---(.(((-(((((((......(((((((((...((((((((.........)-)))))))...)))).)))))((((....))))))))))))))))..))))..))......... ( -31.50, z-score = -3.02, R) >droSec1.super_42 81997 125 + 298440 UAGCGAUGGGUAAA---ACAGA-AACAUCGAUUAUCGAUGCUUGAAAAUUUUGAUGUUUCCGAUUU-AUUAAAAACAUGGAUGUAUCGCGAUACAUCGCCGAUGUUUCUUUCAACUCUUUCUUUUACUUU ....((.((((..(---(.(((-(((((((......(((((((.(...((((((((.........)-)))))))...).)).)))))((((....))))))))))))))))..))))..))......... ( -28.10, z-score = -1.83, R) >droYak2.chr2h_random 646676 113 - 3774259 AAGUGUUGGGUAAA---GCGGA-AACAUCGAACAUCGAUGUUUAAAAAUCUCGAUGUUUCCGAUG--AUUUGAAACAUCGAUGUAUCG----------CCGAUGUUUUUUCCAGCUCUACUUUUAAUAU- ..((((((((((.(---(((((-(((((((.((((((((((((..((((((((.......))).)--)))).))))))))))))....----------.))))))...)))).))).))))..))))))- ( -38.20, z-score = -5.06, R) >droMoj3.scaffold_6473 17351 126 - 16943266 UAGUGGUGGGAACAUUUAUACACAACAUCGAUCAUCGAAAAACAAACAU--CGAUGUUUCUCAUUUUAUUUCAAACAUCGAUGUAUCG--AUAUUACAUCGAUGUUGCGCCCAGCACUACUGGACAGCCU (((((.((((..........(.((((((((((.(((((.......((((--((((((((.............)))))))))))).)))--)).....)))))))))).))))).)))))..((....)). ( -39.12, z-score = -5.16, R) >consensus UAGUGAUGGGUAAA___ACAGA_AACAUCGAACAUCGAUGCUUGAAAAUUUCGAUGUUUCCGAUUU_AUUUAAAACAUCGAUGUAUCGC_AUACAUCGCCGAUGUUUCUUCCAGCUCUAUUUUUUACUUU .((.(.((((................((((((((((((............))))))))..))))........((((((((...................))))))))..)))).).))............ (-12.69 = -12.97 + 0.28)
| Location | 22,471,201 – 22,471,323 |
|---|---|
| Length | 122 |
| Sequences | 5 |
| Columns | 130 |
| Reading direction | reverse |
| Mean pairwise identity | 71.72 |
| Shannon entropy | 0.49452 |
| G+C content | 0.35196 |
| Mean single sequence MFE | -34.26 |
| Consensus MFE | -11.22 |
| Energy contribution | -11.38 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.40 |
| Mean z-score | -3.85 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.996860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
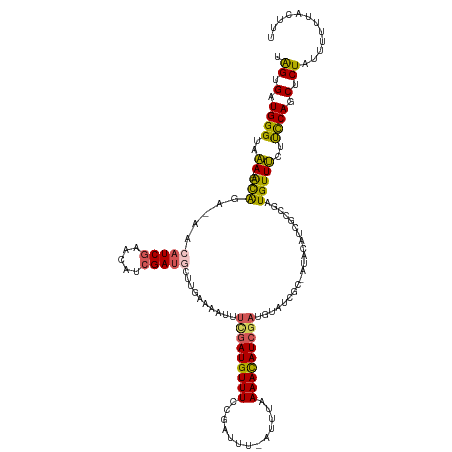
>dm3.chr2L 22471201 122 - 23011544 AAACUACAAAAUAGAGCUGGAAGAAACAUCGGAGAUA----GCGCUACAUAGAUAUUUAAAAAAAAAUCGGAAACAUCGAAAUUUUCGGACAUCGAUGUUAGAUUUU-UCUGU---UUUACCCAUCACCA .......((((((((((((...((....)).....))----))....................((((((...((((((((..((....))..)))))))).))))))-)))))---)))........... ( -20.80, z-score = -0.79, R) >droSim1.chr2h_random 3518 125 + 3178526 AAAGUAAAAGAAAGAGUUGAAAGAAACAUCGGCGAUGUAUCGCGAUACAUCGAUGUUUUUAAU-AAAUCGGAAACAUCAAAAUUUUCAAGCAUCGAUAUUCGAUGUU-UCUGU---UUUACCCAUCGCUA ...(((((((((((..((((.(((((((((...(((((((....))))))))))))))))...-.....(....).))))..)))))((((((((.....)))))))-)...)---)))))......... ( -34.50, z-score = -3.65, R) >droSec1.super_42 81997 125 - 298440 AAAGUAAAAGAAAGAGUUGAAAGAAACAUCGGCGAUGUAUCGCGAUACAUCCAUGUUUUUAAU-AAAUCGGAAACAUCAAAAUUUUCAAGCAUCGAUAAUCGAUGUU-UCUGU---UUUACCCAUCGCUA ...(((((((((((..((((.((((((((..(.(((((((....))))))))))))))))...-.....(....).))))..)))))((((((((.....)))))))-)...)---)))))......... ( -31.50, z-score = -3.28, R) >droYak2.chr2h_random 646676 113 + 3774259 -AUAUUAAAAGUAGAGCUGGAAAAAACAUCGG----------CGAUACAUCGAUGUUUCAAAU--CAUCGGAAACAUCGAGAUUUUUAAACAUCGAUGUUCGAUGUU-UCCGC---UUUACCCAACACUU -.........(((((((.((((...(((((((----------....((((((((((((.((((--(.((((.....)))))))))..))))))))))))))))))))-)))))---)))))......... ( -40.20, z-score = -6.76, R) >droMoj3.scaffold_6473 17351 126 + 16943266 AGGCUGUCCAGUAGUGCUGGGCGCAACAUCGAUGUAAUAU--CGAUACAUCGAUGUUUGAAAUAAAAUGAGAAACAUCG--AUGUUUGUUUUUCGAUGAUCGAUGUUGUGUAUAAAUGUUCCCACCACUA .((.((.((((.....))))(((((((((((((....(((--(((.((((((((((((.............))))))))--)))).......)))))))))))))))))))...........)))).... ( -44.32, z-score = -4.78, R) >consensus AAACUAAAAAAUAGAGCUGGAAGAAACAUCGGCGAUAUAU_GCGAUACAUCGAUGUUUUAAAU_AAAUCGGAAACAUCGAAAUUUUCAAACAUCGAUGUUCGAUGUU_UCUGU___UUUACCCAUCACUA .................((((((....((((...........)))).....((((((((...........))))))))....))))))(((((((.....)))))))....................... (-11.22 = -11.38 + 0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:58:21 2011