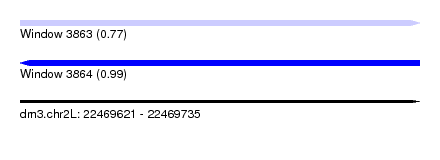
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 22,469,621 – 22,469,735 |
| Length | 114 |
| Max. P | 0.992934 |

| Location | 22,469,621 – 22,469,735 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 84.03 |
| Shannon entropy | 0.26406 |
| G+C content | 0.53806 |
| Mean single sequence MFE | -35.75 |
| Consensus MFE | -29.36 |
| Energy contribution | -29.43 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.773534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
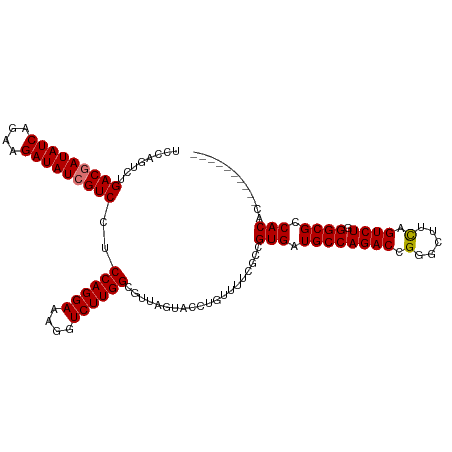
>dm3.chr2L 22469621 114 + 23011544 UCCAGUCUGACAAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACCUGUUUUCGCCGUGAUGCCAGACCGGGCUUCAGUCUGGGCGCCACACUACUACUGCC (((((.((((..(((((....))))).((((((...))..(((((.(((((((................)))))))))))))))).)))).))))).................. ( -31.59, z-score = 0.22, R) >droSim1.chr2h_random 2495535 104 - 3178526 GCCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACCUGUUUUCGCCGUGAUGCCAGACCGGGCUUCAGUCUGGGCGCCACAC---------- (((.(((((((((((((....))))))))).....((....(((.(((((..((......))..))))).))).))))))(((((....)))))))).......---------- ( -37.10, z-score = -1.74, R) >droSec1.super_0 20531174 97 - 21120651 UCCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGG------UACUUGUUUUCGCCGUGAUGCCAGACCGGGCUUCAGUCUGGGCGCCACA----------- (((((.(((((((((((....)))))))(((....)))..(((((.((------((.(..(.......)..)))))))))).....)))).))))).......----------- ( -32.50, z-score = -1.17, R) >droEre2.scaffold_4929 21984527 107 + 26641161 -----UCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACUUGCUUACGCCGUGUUGCCAGACUGGGCUUUAGUCUGGGCGCCACAUCUACUCCC-- -----...(((((((((....))))))))).....(((.((((...(((((.(((....))).)))))(((.((((((((((.....)))))).)))).)))))))..))).-- ( -41.80, z-score = -3.38, R) >consensus UCCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACCUGUUUUCGCCGUGAUGCCAGACCGGGCUUCAGUCUGGGCGCCACAC__________ ........(((((((((....)))))))))..((((((....))))))....................(((.((((((((.(.....).)))).)))).)))............ (-29.36 = -29.43 + 0.06)

| Location | 22,469,621 – 22,469,735 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 84.03 |
| Shannon entropy | 0.26406 |
| G+C content | 0.53806 |
| Mean single sequence MFE | -39.31 |
| Consensus MFE | -34.62 |
| Energy contribution | -34.99 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.992934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
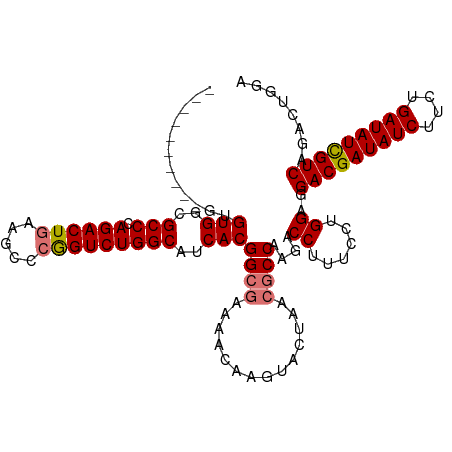
>dm3.chr2L 22469621 114 - 23011544 GGCAGUAGUAGUGUGGCGCCCAGACUGAAGCCCGGUCUGGCAUCACGGCGAAAACAGGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUUGUCAGACUGGA (((..(((((.(((..(((((((((((.....))))))).......))))...)))..)))))..)))........(((.(...(((((((((....)))))))))...).))) ( -41.51, z-score = -1.97, R) >droSim1.chr2h_random 2495535 104 + 3178526 ----------GUGUGGCGCCCAGACUGAAGCCCGGUCUGGCAUCACGGCGAAAACAGGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGC ----------..(((..(((.((((((.....)))))))))..)))((((..............))))..........(..(..(((((((((....)))))))))...)..). ( -38.44, z-score = -2.32, R) >droSec1.super_0 20531174 97 + 21120651 -----------UGUGGCGCCCAGACUGAAGCCCGGUCUGGCAUCACGGCGAAAACAAGUA------CCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGA -----------.(((..(((.((((((.....)))))))))..)))..............------(((...((......))..(((((((((....)))))))))....))). ( -35.40, z-score = -2.44, R) >droEre2.scaffold_4929 21984527 107 - 26641161 --GGGAGUAGAUGUGGCGCCCAGACUAAAGCCCAGUCUGGCAACACGGCGUAAGCAAGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGA----- --(((((.((..(((..(((.(((((.......))))))))..)))(((((.((......)).))))).....)))))))....(((((((((....)))))))))...----- ( -41.90, z-score = -3.62, R) >consensus __________GUGUGGCGCCCAGACUGAAGCCCGGUCUGGCAUCACGGCGAAAACAAGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGA ............(((..(((.((((((.....)))))))))..)))((((..............))))....((......))..(((((((((....)))))))))........ (-34.62 = -34.99 + 0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:58:19 2011