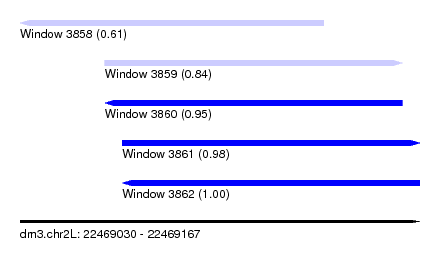
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 22,469,030 – 22,469,167 |
| Length | 137 |
| Max. P | 0.998733 |

| Location | 22,469,030 – 22,469,134 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 69.99 |
| Shannon entropy | 0.57081 |
| G+C content | 0.46328 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -8.32 |
| Energy contribution | -9.97 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.610005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 22469030 104 - 23011544 UAACG-CCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGACACACUGGAACUUCAGCUCCG-CGCGCAACAAGCUUACGGCA--UAAAACAACA--- ....(-((..........(((((((((((((((....)))))))))...(..(.....)..)..)))))).....-.((.......))....))).--..........--- ( -29.00, z-score = -3.08, R) >droSec1.super_394 10130 105 - 13123 UAACG-CCAAGACCUUUCUUGGAGGACGAUAUCUUCUGAUAUCGCCAGACUGGGCUUACGGGAACUUCAGCUCCGCCGCGCAACAAGCUUACGGCA--UAAAACAACA--- .....-((((((....)))))).((.(((((((....))))))))).....(((((...(....)...))).))((((.((.....))...)))).--..........--- ( -31.90, z-score = -2.97, R) >droEre2.scaffold_4929 4553253 105 + 26641161 UAACG-CCAAGACCUUUCCUGGAGGACGAUAUCUUCUAAUAUCGUCAGACUGGUCACUCUCG-ACUUCAGCCUUGCCGCGCAACAAGCUUACGGCA--UCAAAUUACCU-- .....-............((((((((((((((......)))))))).((....)).......-.))))))...(((((.((.....))...)))))--...........-- ( -24.70, z-score = -1.66, R) >droYak2.chr2h_random 1064689 104 + 3774259 UAACG-CCAAGAACUUUCCUGGAAGACGAUAGCUUCUGAUAUCGUCAGACUGGCAACUCUGG-ACUUCAACUCGGCCGAGCAACAAGCUUACGGCU--UCAAAUCAAC--- ....(-(((....((.....))..(((((((........)))))))....))))........-..........((((((((.....)))..)))))--..........--- ( -23.90, z-score = -0.97, R) >droSim1.chrU 10451054 106 + 15797150 UAACG-CCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGCCACACUGGAACUUCAACGCCG-CGCGCAACAAGCUUACUUUAAUCUAAACAAUA--- ....(-(((...((......))..(((((((((....)))))))))....)))).....((((........((..-.))((.....)).........)))).......--- ( -23.00, z-score = -1.67, R) >droMoj3.scaffold_4024 8657 104 - 9767 UCUCGAUCGAUACUAUCGAUUAAACACGAUCUCUCAAUUUACCAUC--AUUAGUAAUAAGAAUAUAUAUACA----CACUUCUUAAGCUU-CAACGCGUCACAUCACACGC ....(((((((...)))))))......((.........((((....--....))))((((((..........----...))))))....)-)...((((........)))) ( -9.72, z-score = -0.43, R) >consensus UAACG_CCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGCCACACUGGAACUUCAACUCCG_CGCGCAACAAGCUUACGGCA__UCAAACAACA___ ...................((((((((((((((....)))))))))...((((.....))))..))))).......................................... ( -8.32 = -9.97 + 1.64)

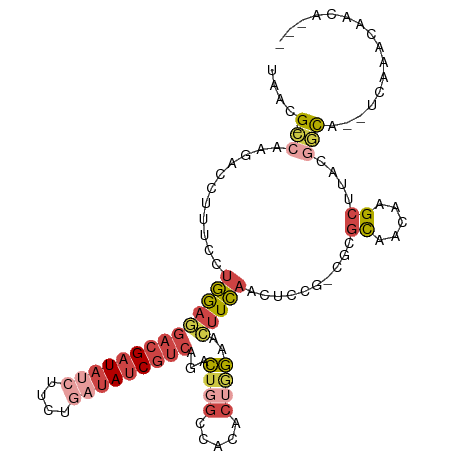
| Location | 22,469,059 – 22,469,161 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 72.92 |
| Shannon entropy | 0.52901 |
| G+C content | 0.48072 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -16.32 |
| Energy contribution | -17.02 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.836910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 22469059 102 + 23011544 GCGGAGCUGAAGUUCCAGUGUGUCCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACCUGUUUUCGCCGUGAUGCCAGAC ..(((((....)))))..........(((((((((((((....))))))))).....((....(((.(((((..((......))..))))).))).)))))) ( -32.50, z-score = -1.73, R) >droSec1.super_394 10160 102 + 13123 GCGGAGCUGAAGUUCCCGUAAGCCCAGUCUGGCGAUAUCAGAAGAUAUCGUCCUCCAAGAAAGGUCUUGGCGUUAGUACCUGUUUUCGCCGUGAUGCCAGAC (((((((....))).)))).......(((((((((((((....))))))..............(((.(((((..((......))..))))).)))))))))) ( -34.30, z-score = -2.65, R) >droEre2.scaffold_4929 4553284 101 - 26641161 GCAAGGCUGAAGU-CGAGAGUGACCAGUCUGACGAUAUUAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACUUGUUUACGCCGUGUUGCCAGAC ((((.((......-.......((((..((((((((((((....))))))))).....)))..))))..(((((............))))))).))))..... ( -31.50, z-score = -1.75, R) >droYak2.chr2h_random 1064719 101 - 3774259 GCCGAGUUGAAGU-CCAGAGUUGCCAGUCUGACGAUAUCAGAAGCUAUCGUCUUCCAGGAAAGUUCUUGGCGUUAGUACUAGCUUUCGCCGUGUUGCCAGAC ((.(..(((....-.)))..).))..(((((.(((((((..((((((..((((.((((((....))))))....)).))))))))..)..)))))).))))) ( -24.90, z-score = 0.15, R) >droSim1.chrU 10451085 102 - 15797150 GCGGCGUUGAAGUUCCAGUGUGGCCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACCUGUUUUCGCCGUGAUGCCAGAC ..((((((.............((((..((((((((((((....))))))))).....)))..))))..((((..((......))..))))..)))))).... ( -33.60, z-score = -1.74, R) >droMoj3.scaffold_4024 8694 85 + 9767 -------AUAUAUAUUCUUAUUACUAAU--GAUGGUAAAUUGAGAGAUCGUGUUUAAUCGAUAGUAUCGA--UCGAGAAUAAU------CGUGAUGCCAGAU -------.......((((((((((((..--..))))))..))))))((((........)))).((((((.--.(((......)------))))))))..... ( -15.40, z-score = 0.22, R) >consensus GCGGAGCUGAAGUUCCAGUGUGACCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACCUGUUUUCGCCGUGAUGCCAGAC ..........................(((((((((((((....)))))))))..........(((..(((((..((......))..)))))....))))))) (-16.32 = -17.02 + 0.70)

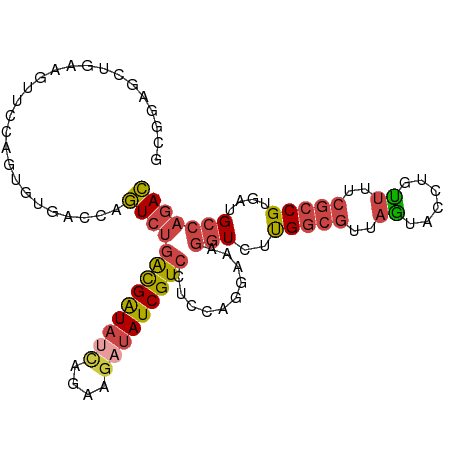
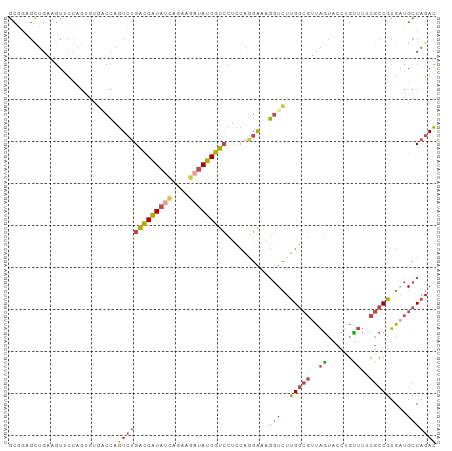
| Location | 22,469,059 – 22,469,161 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 72.92 |
| Shannon entropy | 0.52901 |
| G+C content | 0.48072 |
| Mean single sequence MFE | -26.74 |
| Consensus MFE | -15.57 |
| Energy contribution | -16.91 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.953693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 22469059 102 - 23011544 GUCUGGCAUCACGGCGAAAACAGGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGACACACUGGAACUUCAGCUCCGC (((..((.((.(((.(((...((((.((.......)))))))))))))).(((((((((....))))))))).).)..))).....(((.(....).))).. ( -30.70, z-score = -2.00, R) >droSec1.super_394 10160 102 - 13123 GUCUGGCAUCACGGCGAAAACAGGUACUAACGCCAAGACCUUUCUUGGAGGACGAUAUCUUCUGAUAUCGCCAGACUGGGCUUACGGGAACUUCAGCUCCGC (((((((.....((((..............))))....((((.....))))..((((((....))))))))))))).(((((...(....)...))).)).. ( -34.64, z-score = -2.78, R) >droEre2.scaffold_4929 4553284 101 + 26641161 GUCUGGCAACACGGCGUAAACAAGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUAAUAUCGUCAGACUGGUCACUCUCG-ACUUCAGCCUUGC (((((....)..(((((............))))).)))).....((((((((((((((......)))))))).((....)).......-.))))))...... ( -28.10, z-score = -2.03, R) >droYak2.chr2h_random 1064719 101 + 3774259 GUCUGGCAACACGGCGAAAGCUAGUACUAACGCCAAGAACUUUCCUGGAAGACGAUAGCUUCUGAUAUCGUCAGACUGGCAACUCUGG-ACUUCAACUCGGC (((((....)).)))....((((((.((....(((.((....)).)))..(((((((........)))))))))))))))....((((-........)))). ( -27.00, z-score = -1.04, R) >droSim1.chrU 10451085 102 + 15797150 GUCUGGCAUCACGGCGAAAACAGGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGCCACACUGGAACUUCAACGCCGC ...........(((((.....((((.((.......))))))....((((((((((((((....)))))))))...(..(.....)..)..))))).))))). ( -31.50, z-score = -2.35, R) >droMoj3.scaffold_4024 8694 85 - 9767 AUCUGGCAUCACG------AUUAUUCUCGA--UCGAUACUAUCGAUUAAACACGAUCUCUCAAUUUACCAUC--AUUAGUAAUAAGAAUAUAUAU------- .............------..((((((.((--(((((...))))))).................((((....--....))))..)))))).....------- ( -8.50, z-score = 0.61, R) >consensus GUCUGGCAUCACGGCGAAAACAAGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGCCACACUGGAACUUCAGCUCCGC (((((((.....((((..............))))...................((((((....))))))))))))).......................... (-15.57 = -16.91 + 1.33)
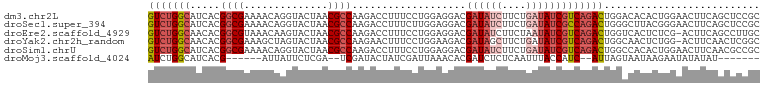
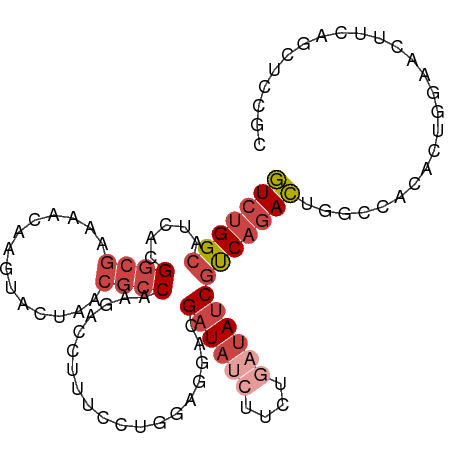
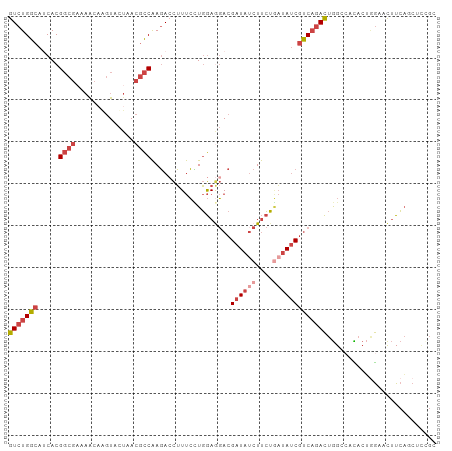
| Location | 22,469,065 – 22,469,167 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 73.71 |
| Shannon entropy | 0.50864 |
| G+C content | 0.47804 |
| Mean single sequence MFE | -31.52 |
| Consensus MFE | -19.18 |
| Energy contribution | -20.22 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.982600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
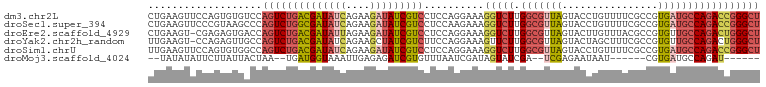
>dm3.chr2L 22469065 102 + 23011544 CUGAAGUUCCAGUGUGUCCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACCUGUUUUCGCCGUGAUGCCAGACCGGGCU (((......)))...((((.(((((((((((((....))))))))).....((....(((.(((((..((......))..))))).))).)))))).)))). ( -34.40, z-score = -2.20, R) >droSec1.super_394 10166 102 + 13123 CUGAAGUUCCCGUAAGCCCAGUCUGGCGAUAUCAGAAGAUAUCGUCCUCCAAGAAAGGUCUUGGCGUUAGUACCUGUUUUCGCCGUGAUGCCAGACCGGGCU ..............(((((.(((((((((((((....))))))..............(((.(((((..((......))..))))).)))))))))).))))) ( -39.40, z-score = -4.09, R) >droEre2.scaffold_4929 4553290 101 - 26641161 CUGAAGU-CGAGAGUGACCAGUCUGACGAUAUUAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACUUGUUUACGCCGUGUUGCCAGACUGGGCU .......-.........((((((((.((((((...(((((....(((....)))...)))))(((((............))))))))))).))))))))... ( -33.90, z-score = -2.06, R) >droYak2.chr2h_random 1064725 101 - 3774259 UUGAAGU-CCAGAGUUGCCAGUCUGACGAUAUCAGAAGCUAUCGUCUUCCAGGAAAGUUCUUGGCGUUAGUACUAGCUUUCGCCGUGUUGCCAGACUGGGCU .......-.........((((((((.(((((((..((((((..((((.((((((....))))))....)).))))))))..)..)))))).))))))))... ( -32.30, z-score = -1.41, R) >droSim1.chrU 10451091 102 - 15797150 UUGAAGUUCCAGUGUGGCCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACCUGUUUUCGCCGUGAUGCCAGACCGGGCU ....(((.((.((.((((......(((((((((....)))))))))...........(((.(((((..((......))..))))).))))))).)).))))) ( -33.70, z-score = -1.66, R) >droMoj3.scaffold_4024 8695 84 + 9767 --UAUAUAUUCUUAUUACUAA--UGAUGGUAAAUUGAGAGAUCGUGUUUAAUCGAUAGUAUCGA--UCGAGAAUAAU------CGUGAUGCCAGAU------ --......((((((((((((.--...))))))..))))))((((........)))).((((((.--.(((......)------)))))))).....------ ( -15.40, z-score = 0.10, R) >consensus CUGAAGUUCCAGUGUGACCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACCUGUUUUCGCCGUGAUGCCAGACCGGGCU .................((.(((((((((((((....)))))))))..........(((..(((((..((......))..)))))....))))))).))... (-19.18 = -20.22 + 1.03)
| Location | 22,469,065 – 22,469,167 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 73.71 |
| Shannon entropy | 0.50864 |
| G+C content | 0.47804 |
| Mean single sequence MFE | -30.75 |
| Consensus MFE | -21.17 |
| Energy contribution | -22.84 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.47 |
| SVM RNA-class probability | 0.998733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
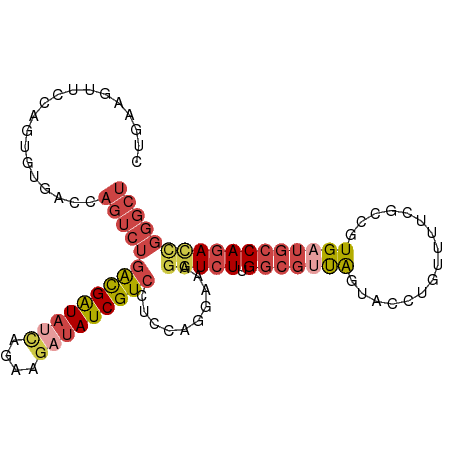
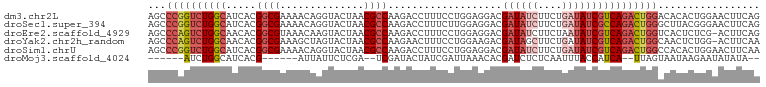
>dm3.chr2L 22469065 102 - 23011544 AGCCCGGUCUGGCAUCACGGCGAAAACAGGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGACACACUGGAACUUCAG .(.((((((((((.....((((..............))))....((((.....))))..((((((....)))))))))))))))).)...((((....)))) ( -34.34, z-score = -2.83, R) >droSec1.super_394 10166 102 - 13123 AGCCCGGUCUGGCAUCACGGCGAAAACAGGUACUAACGCCAAGACCUUUCUUGGAGGACGAUAUCUUCUGAUAUCGCCAGACUGGGCUUACGGGAACUUCAG (((((((((((((.....((((..............))))....((((.....))))..((((((....))))))))))))))))))).............. ( -43.04, z-score = -5.10, R) >droEre2.scaffold_4929 4553290 101 + 26641161 AGCCCAGUCUGGCAACACGGCGUAAACAAGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUAAUAUCGUCAGACUGGUCACUCUCG-ACUUCAG .(.((((((((((.....(((((............)))))....((((.....))))..(((((......))))))))))))))).).......-....... ( -31.90, z-score = -3.26, R) >droYak2.chr2h_random 1064725 101 + 3774259 AGCCCAGUCUGGCAACACGGCGAAAGCUAGUACUAACGCCAAGAACUUUCCUGGAAGACGAUAGCUUCUGAUAUCGUCAGACUGGCAACUCUGG-ACUUCAA .(((.((((((((.....(((....))).((....)))))................(((((((........)))))))))))))))........-....... ( -31.20, z-score = -2.07, R) >droSim1.chrU 10451091 102 + 15797150 AGCCCGGUCUGGCAUCACGGCGAAAACAGGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGCCACACUGGAACUUCAA ...(((((.((((.((.(((.(((...((((.((.......)))))))))))))).(((((((((....)))))))))......)))).)))))........ ( -35.50, z-score = -3.02, R) >droMoj3.scaffold_4024 8695 84 - 9767 ------AUCUGGCAUCACG------AUUAUUCUCGA--UCGAUACUAUCGAUUAAACACGAUCUCUCAAUUUACCAUCA--UUAGUAAUAAGAAUAUAUA-- ------.............------..((((((.((--(((((...))))))).................((((.....--...))))..))))))....-- ( -8.50, z-score = 0.55, R) >consensus AGCCCGGUCUGGCAUCACGGCGAAAACAAGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGCCACACUGGAACUUCAG ...((((((((((.....((((..............))))...................((((((....))))))))))))))))................. (-21.17 = -22.84 + 1.67)
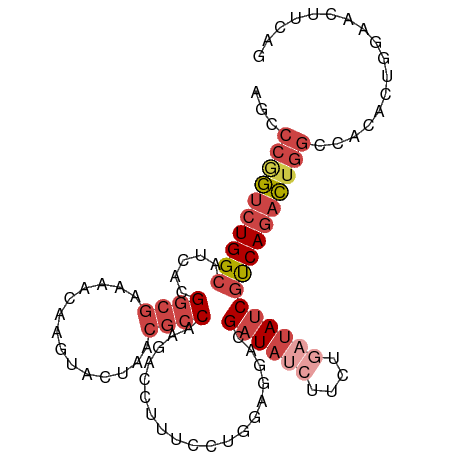
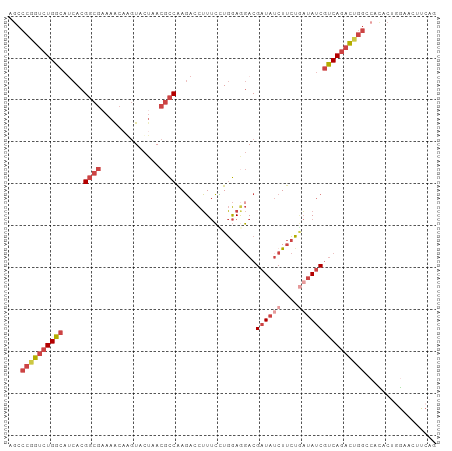
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:58:18 2011