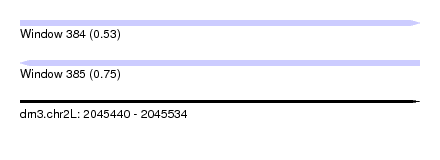
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,045,440 – 2,045,534 |
| Length | 94 |
| Max. P | 0.750964 |

| Location | 2,045,440 – 2,045,534 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 64.64 |
| Shannon entropy | 0.64286 |
| G+C content | 0.49443 |
| Mean single sequence MFE | -31.23 |
| Consensus MFE | -9.42 |
| Energy contribution | -9.00 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.534281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
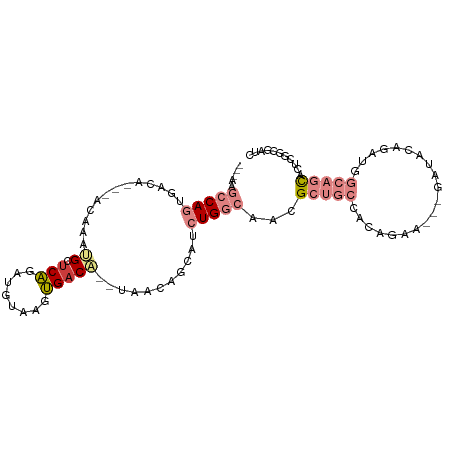
>dm3.chr2L 2045440 94 + 23011544 GAUCGCCGAGUGCUGCCACUUGUAUCCUCUUCUGUGGCAGCGUUGCCAGAUGCUGUCA--UGUCACUUACAUCUGAGCAUUUGU---UGUCACUGGCUU--- ....(((.(((((((((((..............))))))))..(((((((((..((..--....))...)))))).))).....---....))))))..--- ( -32.54, z-score = -2.16, R) >droPer1.super_8 21106 102 + 3966273 GAACGGCGAGUAUUGUUAUCGAUAGCAGGCGAUCUUUUGCCAUUCACAGGGAUGAUCAGCUGUCGACAGCUGUCGAUCGUUUGUAUUUGUUGUUGGUGGUGC .((((((((((((....(((((((((.(((((....)))))..((((((.(.....)..)))).))..))))))))).....))))))))))))........ ( -35.20, z-score = -2.26, R) >dp4.chr4_group3 8838820 102 + 11692001 GAACGGCGAGUAUUGUUAUCGAUAGCAGGCGAUCUUUUGCCAUUCACAGGGAUGAUCAGCUGUCGACAGCUGUCGAUCGUUUGUAUUUGUUGUUGGUGGUGC .((((((((((((....(((((((((.(((((....)))))..((((((.(.....)..)))).))..))))))))).....))))))))))))........ ( -35.20, z-score = -2.26, R) >droSim1.chr2L 2015341 91 + 22036055 GAUCUCCAAGUGCUGCCAUCUGUAUC---UUCUGUGGCAGCGUUGCCAGAUGCUGUUA--UGUCACCUACAUCUGUGCAUUUGU---UGUCACUGGCUU--- ..............((((........---....(((((((((.(((((((((..((..--....))...)))))).)))..)))---))))))))))..--- ( -26.00, z-score = -1.15, R) >droSec1.super_14 1992161 91 + 2068291 GAUCUCAAAGUGCUGCCAUCUGUAUC---UUCUGUGGCAGCGUUGCCAGAUGCUGUUA--UGUCACUUACAUCUGAGCAUUUGU---UGUCACUGGCUU--- (((..((((..((((((((..(....---..).))))))))..(((((((((..((..--....))...)))))).))))))).---.)))........--- ( -28.50, z-score = -1.80, R) >droYak2.chr2L 2031064 86 + 22324452 GAUCGCCAAGUGCUGCC-----CAUC---UUUUGCGGCAGCGUUGCCAGAUGCUGUUA--CGUCAUUGACAUCUGAGCGUUUGU---UGUCACUGGCUU--- ....((((.((((((((-----((..---...)).))))))...(((((((((.....--.......).)))))).))......---....))))))..--- ( -29.10, z-score = -1.35, R) >droEre2.scaffold_4929 2093121 94 + 26641161 GAUCGCCCAGUGCUGCCACUUCUAUC---UGCUGAUGCAGCGUUGCCAGAUGCUGUUA--UGUCAUUGACAGCUGAGCAUUUCU---UGUCACUGGCGGCUU ....(((....(((((....((....---....)).)))))...(((((..(((((((--......)))))))(((.((.....---))))))))))))).. ( -32.10, z-score = -1.05, R) >consensus GAUCGCCAAGUGCUGCCAUCUGUAUC___UUCUGUGGCAGCGUUGCCAGAUGCUGUUA__UGUCACUUACAUCUGAGCAUUUGU___UGUCACUGGCUU___ ........((((((((....................))))))))(((((..((.......(((((........))).)).........))..)))))..... ( -9.42 = -9.00 + -0.42)

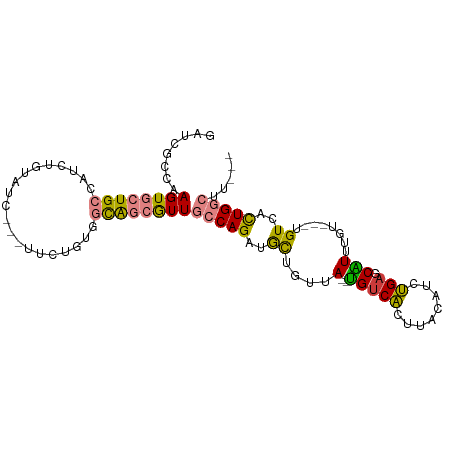
| Location | 2,045,440 – 2,045,534 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 64.64 |
| Shannon entropy | 0.64286 |
| G+C content | 0.49443 |
| Mean single sequence MFE | -27.66 |
| Consensus MFE | -7.09 |
| Energy contribution | -8.74 |
| Covariance contribution | 1.66 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.750964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2045440 94 - 23011544 ---AAGCCAGUGACA---ACAAAUGCUCAGAUGUAAGUGACA--UGACAGCAUCUGGCAACGCUGCCACAGAAGAGGAUACAAGUGGCAGCACUCGGCGAUC ---..((((((....---.....(((.(((((((..((....--..)).))))))))))..((((((((..............))))))))))).))).... ( -34.54, z-score = -3.38, R) >droPer1.super_8 21106 102 - 3966273 GCACCACCAACAACAAAUACAAACGAUCGACAGCUGUCGACAGCUGAUCAUCCCUGUGAAUGGCAAAAGAUCGCCUGCUAUCGAUAACAAUACUCGCCGUUC ..................(((...(((.((((((((....)))))).)))))..)))(((((((....(((.(....).)))((.........))))))))) ( -22.00, z-score = -1.89, R) >dp4.chr4_group3 8838820 102 - 11692001 GCACCACCAACAACAAAUACAAACGAUCGACAGCUGUCGACAGCUGAUCAUCCCUGUGAAUGGCAAAAGAUCGCCUGCUAUCGAUAACAAUACUCGCCGUUC ..................(((...(((.((((((((....)))))).)))))..)))(((((((....(((.(....).)))((.........))))))))) ( -22.00, z-score = -1.89, R) >droSim1.chr2L 2015341 91 - 22036055 ---AAGCCAGUGACA---ACAAAUGCACAGAUGUAGGUGACA--UAACAGCAUCUGGCAACGCUGCCACAGAA---GAUACAGAUGGCAGCACUUGGAGAUC ---...(((((....---.....(((.(((((((..((....--..)).))))))))))..((((((((....---......).))))))))).)))..... ( -28.60, z-score = -2.88, R) >droSec1.super_14 1992161 91 - 2068291 ---AAGCCAGUGACA---ACAAAUGCUCAGAUGUAAGUGACA--UAACAGCAUCUGGCAACGCUGCCACAGAA---GAUACAGAUGGCAGCACUUUGAGAUC ---..(((((.....---....(((((...((((.....)))--)...))))))))))...((((((((....---......).)))))))........... ( -26.10, z-score = -2.05, R) >droYak2.chr2L 2031064 86 - 22324452 ---AAGCCAGUGACA---ACAAACGCUCAGAUGUCAAUGACG--UAACAGCAUCUGGCAACGCUGCCGCAAAA---GAUG-----GGCAGCACUUGGCGAUC ---..((((((....---......((.(((((((...((...--...)))))))))))...((((((.((...---..))-----)))))))).)))).... ( -30.50, z-score = -2.63, R) >droEre2.scaffold_4929 2093121 94 - 26641161 AAGCCGCCAGUGACA---AGAAAUGCUCAGCUGUCAAUGACA--UAACAGCAUCUGGCAACGCUGCAUCAGCA---GAUAGAAGUGGCAGCACUGGGCGAUC ..(((((((((((((---.....)).)))(((((........--..)))))..)))))...(((((....((.---.......)).)))))....))).... ( -29.90, z-score = -0.85, R) >consensus ___AAGCCAGUGACA___ACAAAUGCUCAGAUGUAAGUGACA__UAACAGCAUCUGGCAACGCUGCCACAGAA___GAUACAGAUGGCAGCACUCGGCGAUC .....(((((.............((.(((........)))))...........)))))...(((((....................)))))........... ( -7.09 = -8.74 + 1.66)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:10:00 2011