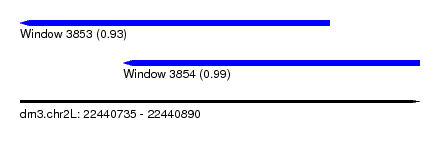
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 22,440,735 – 22,440,890 |
| Length | 155 |
| Max. P | 0.991359 |

| Location | 22,440,735 – 22,440,855 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.27 |
| Shannon entropy | 0.43365 |
| G+C content | 0.47328 |
| Mean single sequence MFE | -37.34 |
| Consensus MFE | -19.82 |
| Energy contribution | -19.86 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 22440735 120 - 23011544 UCCCUCAAUCGAUGCAUCUGGUCGUUACAGCACUCUUGCGACACUUUUACUGGUGUCGGUGUGUCUUGAAGCCUUAGAAAGCUAUCCGUACGAAUUCUAGGUAGUGCAUGUGGUGGUAUU ..((((((..((((((((...........(((....)))((((((......)))))))))))))))))).(((.((....((((.((............))))))...)).))))).... ( -29.00, z-score = 0.65, R) >droSim1.chr2h_random 1259888 105 - 3178526 UCCCUCAAUCGAUGCAUCUAGGCGUUAAAGCACUCUUGUGACACUUGUUCUGGUGUCGGGGUGUCUUGAUGCCUUAGAAAGCUAUCCGUAGGGUUUCUAGGUAGU--------------- .((((..((.(((((.((((((((((((..((((((...((((((......))))))))))))..)))))))).))))..)).))).))))))............--------------- ( -38.60, z-score = -3.52, R) >droSec1.super_42 48741 105 - 298440 UCCCUCAAUCGAUGCAUCUAGGCGUUAAAGCACUCUUGUGACACUUGUUCUUGUGUCGGGGUGUCUUGAUGCCUUAGAAAGCUAUACGUAGGGUUUCUAGGUAGU--------------- .((((....((((((.((((((((((((..((((((...(((((........)))))))))))..)))))))).))))..)).)).)).))))............--------------- ( -36.30, z-score = -3.12, R) >droYak2.chr2R 20774275 120 + 21139217 UCCCUUAACCGAUGCAUUUGGCUGUUAAAACAUUCUUGCGACACCUGUUCUGGUGUUGGGGUAUCUUGAUGUCUUAAAAAGCUAUCUGUGCGGAUACUGGGUAGUCCAGGUGGUGGGGCC .((((..(((...((((.(((((.((((.((((((((.(((((((......))))))))))......))))).))))..)))))...))))((((........)))).)))...)))).. ( -38.00, z-score = -1.30, R) >droEre2.scaffold_4784 24376539 118 - 25762168 UCCCUUGACCGGUACAUCUGGCUAUUAAAGCACUCUGACGACACUUUUUCUGUUGUCGGUGUGUUUUAAUGCCUUAAGAAGCUAUCGGUAGGGAUUCUAGG--GUUCAGGUGGCGGUGGC (((((..((((((((.((((((.((((((((((.(((((((((.......))))))))).)))))))))))))...))).).)))))))))))).......--................. ( -44.80, z-score = -3.55, R) >consensus UCCCUCAAUCGAUGCAUCUGGCCGUUAAAGCACUCUUGCGACACUUGUUCUGGUGUCGGGGUGUCUUGAUGCCUUAGAAAGCUAUCCGUAGGGAUUCUAGGUAGU_CA_GUGG_GG____ .((((..((.(((((.(((((.((((((.((((((...(((((((......))))))))))))).)))))).).))))..)).))).))))))........................... (-19.82 = -19.86 + 0.04)


| Location | 22,440,775 – 22,440,890 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.96 |
| Shannon entropy | 0.39320 |
| G+C content | 0.48036 |
| Mean single sequence MFE | -39.06 |
| Consensus MFE | -22.80 |
| Energy contribution | -22.32 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.991359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
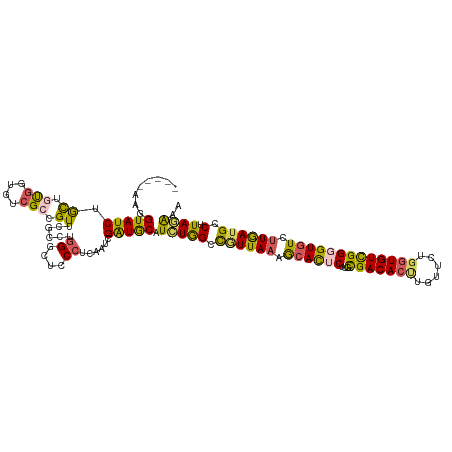
>dm3.chr2L 22440775 115 - 23011544 -----AAGGUAUCUACUGUGGUUUCGCCGUUCGCCUGGGCUCCCUCAAUCGAUGCAUCUGGUCGUUACAGCACUCUUGCGACACUUUUACUGGUGUCGGUGUGUCUUGAAGCCUUAGAAA -----..........(((.(((((((((.........)))..........((((((((...........(((....)))((((((......))))))))))))))..)))))).)))... ( -33.60, z-score = -0.95, R) >droSim1.chr2h_random 1259913 120 - 3178526 GUGGUAUCGUAUCUGCUAUGGUGUCGAAGUUUGCUCGGUCUCCCUCAAUCGAUGCAUCUAGGCGUUAAAGCACUCUUGUGACACUUGUUCUGGUGUCGGGGUGUCUUGAUGCCUUAGAAA .((..((((((.....))))))..)).....((((((((........))))).)))((((((((((((..((((((...((((((......))))))))))))..)))))))).)))).. ( -43.50, z-score = -4.05, R) >droSec1.super_42 48766 120 - 298440 GAGGUAUCGUAUCUGCUAUGAUGUCGAAGUUUGCUCGGGCUCCCUCAAUCGAUGCAUCUAGGCGUUAAAGCACUCUUGUGACACUUGUUCUUGUGUCGGGGUGUCUUGAUGCCUUAGAAA ((((..(((((((......)))).)))(((((....))))).))))..........((((((((((((..((((((...(((((........)))))))))))..)))))))).)))).. ( -42.30, z-score = -3.27, R) >droYak2.chr2R 20774315 115 + 21139217 -----ACGGUAUCUGCUGCGGAGUGGCCGUUCGUCUGGGAUCCCUUAACCGAUGCAUUUGGCUGUUAAAACAUUCUUGCGACACCUGUUCUGGUGUUGGGGUAUCUUGAUGUCUUAAAAA -----(((((.((((...))))...))))).((((.(((((((((........(((...(..(((....)))..).)))((((((......)))))))))).)))))))))......... ( -32.70, z-score = -0.85, R) >droEre2.scaffold_4784 24376577 115 - 25762168 -----GAGGUAUCUAUUGUGGUGUCGCCGUUCGUCUAGGCUCCCUUGACCGGUACAUCUGGCUAUUAAAGCACUCUGACGACACUUUUUCUGUUGUCGGUGUGUUUUAAUGCCUUAAGAA -----......(((.....(((((.((((...(((.(((...))).)))))))))))).(((.((((((((((.(((((((((.......))))))))).)))))))))))))...))). ( -43.20, z-score = -4.97, R) >consensus _____AAGGUAUCUGCUGUGGUGUCGCCGUUCGCCUGGGCUCCCUCAAUCGAUGCAUCUGGCCGUUAAAGCACUCUUGCGACACUUGUUCUGGUGUCGGGGUGUCUUGAUGCCUUAGAAA ........(((((.((.(((....))).))......((....))......))))).(((((.((((((.((((((...(((((((......))))))))))))).)))))).).)))).. (-22.80 = -22.32 + -0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:58:12 2011