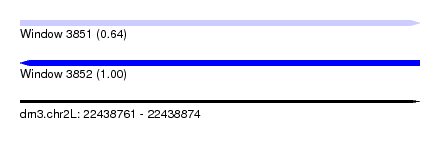
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 22,438,761 – 22,438,874 |
| Length | 113 |
| Max. P | 0.995464 |

| Location | 22,438,761 – 22,438,874 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 84.89 |
| Shannon entropy | 0.26739 |
| G+C content | 0.48190 |
| Mean single sequence MFE | -34.26 |
| Consensus MFE | -23.82 |
| Energy contribution | -24.70 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.640591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 22438761 113 + 23011544 UAAU-UUUAUUUUCGUUAUGUUGGAGAAGUGAGAGGCUUACCAAGAUCCCAGGAACCGAAUACG-CGAGCUUAGGCUGGCAAACUGGUGCCUGAACAU-GGCUGCCUCUCCCUC-GC ....-....((((((......))))))((.(((((((...(((.(....((((.((((.....(-(.(((....))).))....)))).))))..).)-))..))))))).)).-.. ( -35.10, z-score = -1.46, R) >droSim1.chr2h_random 1258893 116 + 3178526 UAAUUUUUAUUCUCUUUAUGUUGAUGAGGUGAGAGGCUUACCAAGAUCCCAUGAACCAAAUACG-CGAGCUUAGGCUUGCAAACUGGUGUCUGAACAUCGGAUGCCUCUCCCUCUGC .........................((((.(((((((..........................(-(((((....))))))...(((((((....)))))))..)))))))))))... ( -37.50, z-score = -3.45, R) >droSec1.super_42 47745 116 + 298440 UAAUAUUUAUUCUCUUUAUGUUGAUGAGGUGAGAGGCUUACCAAGAUCCCAUGAACCAAAUACG-CGAGCUUAGGCUUGCAAACUGGUGUCUGAACAUCGGAUGCCUCUCCCUCUGC .........................((((.(((((((..........................(-(((((....))))))...(((((((....)))))))..)))))))))))... ( -37.50, z-score = -3.48, R) >droEre2.scaffold_4784 24210917 112 + 25762168 ---UAAAUAUUUUCGUUAUGUUGGAGAGGUAAGAGGCUUACCAAGAUCCCACGA-CUAAAUACGACGAGCUUACGCUGGCAACCUGGUGCCUGAACAUCGGAUGCCUCUCCCUCGG- ---...............((..(((((((((..((((..((((.(...((.((.-.......))...(((....)))))...).))))))))(.....)...)))))))))..)).- ( -27.50, z-score = -0.15, R) >droYak2.chr2R 20773049 114 - 21139217 UAAAAUUUAUUGUCGUUAUGUUGGGGAGGUAAGGGGCUUACCAAGAUCCCACGACCCAAAAUCGACGAGCUUACGCCGGCAUCCUGGUCCCUGAACAUCGGAUGCCCCUCCCUC--- .........((((((.....(((((..(....((((((.....)).)))).)..)))))...))))))((....)).(((((((.(((........))))))))))........--- ( -33.70, z-score = -0.45, R) >consensus UAAUAUUUAUUCUCGUUAUGUUGGAGAGGUGAGAGGCUUACCAAGAUCCCACGAACCAAAUACG_CGAGCUUAGGCUGGCAAACUGGUGCCUGAACAUCGGAUGCCUCUCCCUC_GC .........................((((.(((((((..........................(.(.(((....))).))...((((((......))))))..)))))))))))... (-23.82 = -24.70 + 0.88)

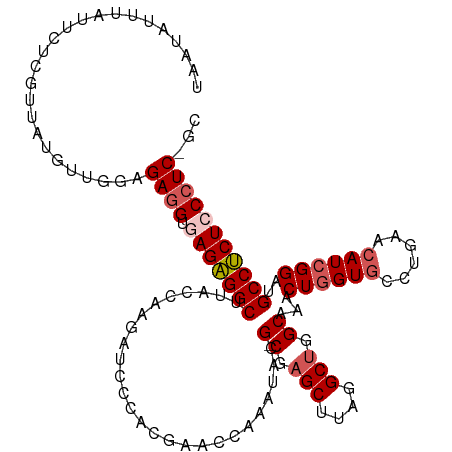
| Location | 22,438,761 – 22,438,874 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 84.89 |
| Shannon entropy | 0.26739 |
| G+C content | 0.48190 |
| Mean single sequence MFE | -40.10 |
| Consensus MFE | -32.10 |
| Energy contribution | -31.98 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.995464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 22438761 113 - 23011544 GC-GAGGGAGAGGCAGCC-AUGUUCAGGCACCAGUUUGCCAGCCUAAGCUCG-CGUAUUCGGUUCCUGGGAUCUUGGUAAGCCUCUCACUUCUCCAACAUAACGAAAAUAAA-AUUA ..-(((((((((((.(((-(..((((((.((((((.(((.(((....))).)-)).))).))).))))))....))))..))))))).))))....................-.... ( -41.00, z-score = -3.40, R) >droSim1.chr2h_random 1258893 116 - 3178526 GCAGAGGGAGAGGCAUCCGAUGUUCAGACACCAGUUUGCAAGCCUAAGCUCG-CGUAUUUGGUUCAUGGGAUCUUGGUAAGCCUCUCACCUCAUCAACAUAAAGAGAAUAAAAAUUA ...(((((((((((..((((.((((.((.(((((..(((.(((....))).)-))...)))))))...)))).))))...))))))).))))......................... ( -40.20, z-score = -3.90, R) >droSec1.super_42 47745 116 - 298440 GCAGAGGGAGAGGCAUCCGAUGUUCAGACACCAGUUUGCAAGCCUAAGCUCG-CGUAUUUGGUUCAUGGGAUCUUGGUAAGCCUCUCACCUCAUCAACAUAAAGAGAAUAAAUAUUA ...(((((((((((..((((.((((.((.(((((..(((.(((....))).)-))...)))))))...)))).))))...))))))).))))......................... ( -40.20, z-score = -3.86, R) >droEre2.scaffold_4784 24210917 112 - 25762168 -CCGAGGGAGAGGCAUCCGAUGUUCAGGCACCAGGUUGCCAGCGUAAGCUCGUCGUAUUUAG-UCGUGGGAUCUUGGUAAGCCUCUUACCUCUCCAACAUAACGAAAAUAUUUA--- -..(((((((((((..((((.((((.((((......))))(((....)))............-.....)))).))))...))))))).))))......................--- ( -35.10, z-score = -1.82, R) >droYak2.chr2R 20773049 114 + 21139217 ---GAGGGAGGGGCAUCCGAUGUUCAGGGACCAGGAUGCCGGCGUAAGCUCGUCGAUUUUGGGUCGUGGGAUCUUGGUAAGCCCCUUACCUCCCCAACAUAACGACAAUAAAUUUUA ---(((((((((((.(((........)))((((((((.(((((....)))...((((.....)))).)).))))))))..))))))).))))......................... ( -44.00, z-score = -2.16, R) >consensus GC_GAGGGAGAGGCAUCCGAUGUUCAGGCACCAGUUUGCCAGCCUAAGCUCG_CGUAUUUGGUUCAUGGGAUCUUGGUAAGCCUCUCACCUCACCAACAUAACGAAAAUAAAUAUUA ...(((((((((((..((((.((((.((.(((((..(((.(((....))).).))...)))))))...)))).))))...))))))).))))......................... (-32.10 = -31.98 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:58:10 2011