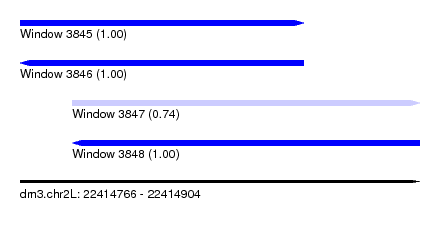
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 22,414,766 – 22,414,904 |
| Length | 138 |
| Max. P | 0.999535 |

| Location | 22,414,766 – 22,414,864 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.18 |
| Shannon entropy | 0.52605 |
| G+C content | 0.60177 |
| Mean single sequence MFE | -46.06 |
| Consensus MFE | -27.04 |
| Energy contribution | -28.72 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.44 |
| SVM RNA-class probability | 0.998666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 22414766 98 + 23011544 --------CUUGGCUUCAGAAU--A------------CCCGGGCUUGAACUUCGGCCGGCGGUAGUGCUCUACUGACUGGGGUCUCGGAGCUCUACUCACUCCACCACCGCCGGCGGCAA --------....(((.......--.------------..((((.......))))(((((((((.(((......(((.((((((......)))))).)))...))).)))))))))))).. ( -33.60, z-score = 0.01, R) >droSim1.chrU 12458103 110 - 15797150 --------UUGGGCUUCAGAAU--ACCACAUACCAUACCCGGGUUUGAACUCCGGCCGGCGGUAGUGCUCUACUGACUGGGGUGUCGGAGCUCUACUCACUCCACUACCGCCGGCGGCAA --------.(((..((((((..--.((.............)).))))))..)))(((((((((((((......(((.((((((......)))))).)))...)))))))))))))..... ( -45.12, z-score = -2.57, R) >droSec1.super_42 16121 110 + 298440 --------UUUGGCUUCAGAAU--ACCACAUACCAUACCUGGGUUUGAACUUCGGCCGGCGGUAGUGCCCUACUGACUGGGGUGUGGGAGCUCUCCUCACUCCACUACCGCCGGCGGCAA --------....((((((((..--.(((...........))).)))))......((((((((((((((((((.....))))).((((((....))).)))..)))))))))))))))).. ( -47.80, z-score = -3.22, R) >droYak2.chr2L 7945874 114 + 22324452 UUUUACCACCGGGAUUAAGUCCCGGUUCCAUACUAUACCUGGGCUUGAACUCCGGCCGGCGUCAGUUCUCU-CCGACAAGGAUGUCGAAGCUG-----ACUCCACUACCGCCGGCGGCAA .....((..((((.((((((((.(((..........))).)))))))).).)))(((((((..(((((.((-.(((((....))))).))..)-----)....)))..)))))))))... ( -43.70, z-score = -2.83, R) >droEre2.scaffold_4929 12734899 115 - 26641161 ----CGGAACCGACGUCGGUUCUGGUUCUGUCCCAC-GUAGGGCUUGAACUCCGGCCGGCGGUAGCACUCUCCUAACUGGGGAGACCGAGCUCUACUCACUCCACUACCGCCGGCCCCGA ----((((((((....))))))))((((.((((...-...))))..))))...((((((((((((..((((((.....))))))...(((.....)))......)))))))))))).... ( -60.10, z-score = -4.99, R) >consensus ________CUGGGCUUCAGAAU__ACCACAUACCAUACCCGGGCUUGAACUCCGGCCGGCGGUAGUGCUCUACUGACUGGGGUGUCGGAGCUCUACUCACUCCACUACCGCCGGCGGCAA .......................................((((.......))))((((((((((((((((((.....))))))...((((.........))))))))))))))))..... (-27.04 = -28.72 + 1.68)

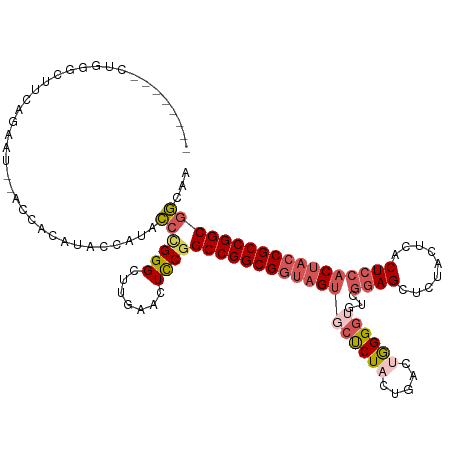
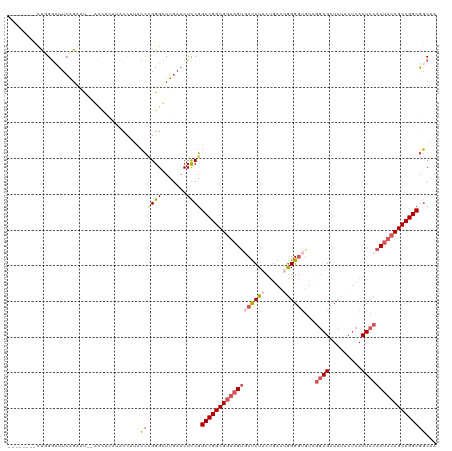
| Location | 22,414,766 – 22,414,864 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.18 |
| Shannon entropy | 0.52605 |
| G+C content | 0.60177 |
| Mean single sequence MFE | -48.00 |
| Consensus MFE | -30.20 |
| Energy contribution | -30.84 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.99 |
| SVM RNA-class probability | 0.999535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 22414766 98 - 23011544 UUGCCGCCGGCGGUGGUGGAGUGAGUAGAGCUCCGAGACCCCAGUCAGUAGAGCACUACCGCCGGCCGAAGUUCAAGCCCGGG------------U--AUUCUGAAGCCAAG-------- .(((((((((((((((((((((.......)))))..(((....)))........))))))))))))((..((....)).))))------------)--).............-------- ( -38.20, z-score = -2.15, R) >droSim1.chrU 12458103 110 + 15797150 UUGCCGCCGGCGGUAGUGGAGUGAGUAGAGCUCCGACACCCCAGUCAGUAGAGCACUACCGCCGGCCGGAGUUCAAACCCGGGUAUGGUAUGUGGU--AUUCUGAAGCCCAA-------- .(((((((((((((((((((((.......)))))(((......)))........))))))))))))(((.((....))))))))).....((.(((--........))))).-------- ( -46.70, z-score = -3.12, R) >droSec1.super_42 16121 110 - 298440 UUGCCGCCGGCGGUAGUGGAGUGAGGAGAGCUCCCACACCCCAGUCAGUAGGGCACUACCGCCGGCCGAAGUUCAAACCCAGGUAUGGUAUGUGGU--AUUCUGAAGCCAAA-------- .(((((((((((((((((..(((.(((....))))))..(((........))))))))))))))))..........((....))..))))..((((--........))))..-------- ( -47.10, z-score = -3.21, R) >droYak2.chr2L 7945874 114 - 22324452 UUGCCGCCGGCGGUAGUGGAGU-----CAGCUUCGACAUCCUUGUCGG-AGAGAACUGACGCCGGCCGGAGUUCAAGCCCAGGUAUAGUAUGGAACCGGGACUUAAUCCCGGUGGUAAAA ((((((((((((((...((.((-----(((((((((((....))))))-))....))))).)).))))......((((((.(((..........)))))).)))....)))))))))).. ( -50.30, z-score = -3.36, R) >droEre2.scaffold_4929 12734899 115 + 26641161 UCGGGGCCGGCGGUAGUGGAGUGAGUAGAGCUCGGUCUCCCCAGUUAGGAGAGUGCUACCGCCGGCCGGAGUUCAAGCCCUAC-GUGGGACAGAACCAGAACCGACGUCGGUUCCG---- ....(((((((((((((.((((.......))))(.(((((.......))))).)))))))))))))).(.((((...(((...-..)))...))))).((((((....))))))..---- ( -57.70, z-score = -3.53, R) >consensus UUGCCGCCGGCGGUAGUGGAGUGAGUAGAGCUCCGACACCCCAGUCAGUAGAGCACUACCGCCGGCCGGAGUUCAAGCCCAGGUAUGGUAUGGAAU__AUUCUGAAGCCAAA________ .(((((((((((((((((((((.......)))))(((......)))........)))))))))))).((.((....)))).))))................................... (-30.20 = -30.84 + 0.64)

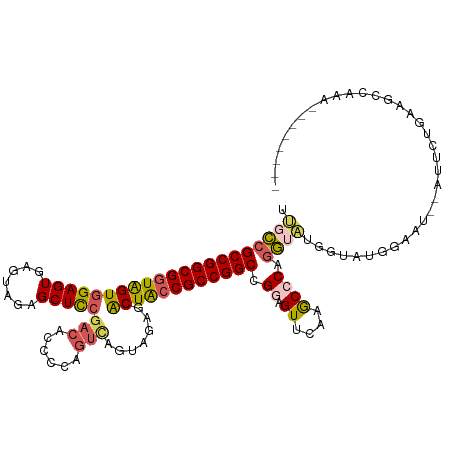
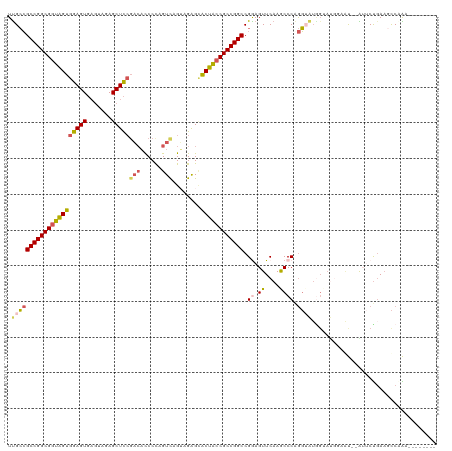
| Location | 22,414,784 – 22,414,904 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.17 |
| Shannon entropy | 0.36176 |
| G+C content | 0.58176 |
| Mean single sequence MFE | -43.40 |
| Consensus MFE | -28.52 |
| Energy contribution | -30.36 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.737379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 22414784 120 + 23011544 GGGCUUGAACUUCGGCCGGCGGUAGUGCUCUACUGACUGGGGUCUCGGAGCUCUACUCACUCCACCACCGCCGGCGGCAAUGGCACAGCAUCCUGGGGUAACCUUGAACUUCGUUUGAUU ..(((((.....(.(((((((((.(((......(((.((((((......)))))).)))...))).))))))))).)......)).))).((..(((....))).))............. ( -38.00, z-score = 0.60, R) >droSim1.chrU 12458133 120 - 15797150 GGGUUUGAACUCCGGCCGGCGGUAGUGCUCUACUGACUGGGGUGUCGGAGCUCUACUCACUCCACUACCGCCGGCGGCAAUGGCAUAGCAUCCUGGGAUAAACUUGAAUUUCGUCUGAUU (((((((...(((.(((((((((((((......(((.((((((......)))))).)))...))))))))))))).((....))...........))))))))))............... ( -43.20, z-score = -1.41, R) >droSec1.super_42 16151 120 + 298440 GGGUUUGAACUUCGGCCGGCGGUAGUGCCCUACUGACUGGGGUGUGGGAGCUCUCCUCACUCCACUACCGCCGGCGGCAAUGGGAUAGCAUCCUGGGAUAAACUUGAAUUUCGUCUGAUU (((((((..((.(.((((((((((((((((((.....))))).((((((....))).)))..))))))))))))).)....(((((...)))))))..)))))))............... ( -48.90, z-score = -2.29, R) >droYak2.chr2L 7945914 114 + 22324452 GGGCUUGAACUCCGGCCGGCGUCAGUUCUCU-CCGACAAGGAUGUCGAAGCUG-----ACUCCACUACCGCCGGCGGCAAUGGGACAGCAUCCUAGGUUAAACUUGAACUUUGUCUGCCC ((((..........(((((((..(((((.((-.(((((....))))).))..)-----)....)))..)))))))((((((((((.....)))))((((.......))))))))).)))) ( -38.90, z-score = -0.90, R) >droEre2.scaffold_4929 12734934 106 - 26641161 GGGCUUGAACUCCGGCCGGCGGUAGCACUCUCCUAACUGGGGAGACCGAGCUCUACUCACUCCACUACCGCCGGCCCCGAUGGGACAGCAUCCUAGGUUUAACUUG-------------- (((.(((((((..((((((((((((..((((((.....))))))...(((.....)))......))))))))))))....(((((.....))))))))))))))).-------------- ( -48.00, z-score = -4.12, R) >consensus GGGCUUGAACUCCGGCCGGCGGUAGUGCUCUACUGACUGGGGUGUCGGAGCUCUACUCACUCCACUACCGCCGGCGGCAAUGGGACAGCAUCCUGGGAUAAACUUGAACUUCGUCUGAUU (((.(((..((.(.((((((((((((((((((.....))))))...((((.........)))))))))))))))).)....))..)))...))).......................... (-28.52 = -30.36 + 1.84)

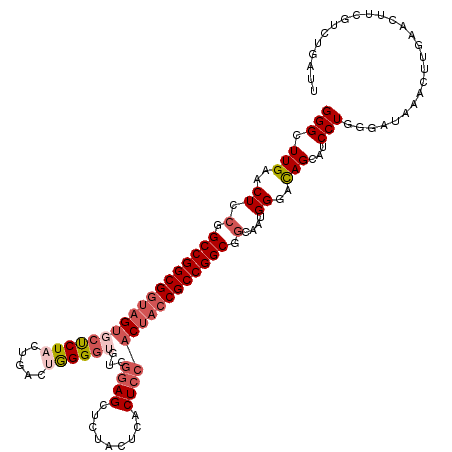
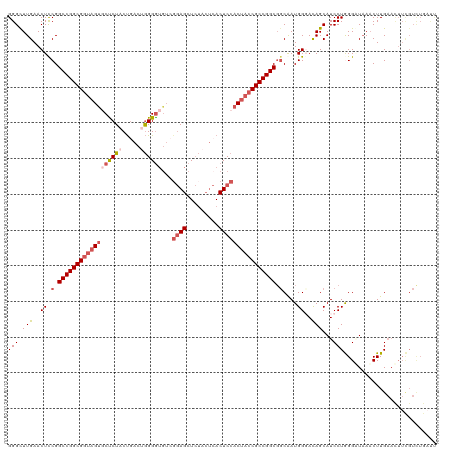
| Location | 22,414,784 – 22,414,904 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.17 |
| Shannon entropy | 0.36176 |
| G+C content | 0.58176 |
| Mean single sequence MFE | -45.14 |
| Consensus MFE | -31.70 |
| Energy contribution | -32.98 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.997407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 22414784 120 - 23011544 AAUCAAACGAAGUUCAAGGUUACCCCAGGAUGCUGUGCCAUUGCCGCCGGCGGUGGUGGAGUGAGUAGAGCUCCGAGACCCCAGUCAGUAGAGCACUACCGCCGGCCGAAGUUCAAGCCC ........(((.(((..(((....((((....))).).....)))(((((((((((((((((.......)))))..(((....)))........)))))))))))).))).)))...... ( -45.50, z-score = -3.05, R) >droSim1.chrU 12458133 120 + 15797150 AAUCAGACGAAAUUCAAGUUUAUCCCAGGAUGCUAUGCCAUUGCCGCCGGCGGUAGUGGAGUGAGUAGAGCUCCGACACCCCAGUCAGUAGAGCACUACCGCCGGCCGGAGUUCAAACCC ........(((.(((.(((..(((....))))))...........(((((((((((((((((.......)))))(((......)))........)))))))))))).))).)))...... ( -41.80, z-score = -3.13, R) >droSec1.super_42 16151 120 - 298440 AAUCAGACGAAAUUCAAGUUUAUCCCAGGAUGCUAUCCCAUUGCCGCCGGCGGUAGUGGAGUGAGGAGAGCUCCCACACCCCAGUCAGUAGGGCACUACCGCCGGCCGAAGUUCAAACCC ........(((.(((.(((..(((....))))))...........(((((((((((((..(((.(((....))))))..(((........)))))))))))))))).))).)))...... ( -46.00, z-score = -3.97, R) >droYak2.chr2L 7945914 114 - 22324452 GGGCAGACAAAGUUCAAGUUUAACCUAGGAUGCUGUCCCAUUGCCGCCGGCGGUAGUGGAGU-----CAGCUUCGACAUCCUUGUCGG-AGAGAACUGACGCCGGCCGGAGUUCAAGCCC .(((.((((..((((.((......)).))))..))))(((((((((....))))))))).((-----(((((((((((....))))))-))....))))))))(((..(....)..))). ( -47.80, z-score = -3.06, R) >droEre2.scaffold_4929 12734934 106 + 26641161 --------------CAAGUUAAACCUAGGAUGCUGUCCCAUCGGGGCCGGCGGUAGUGGAGUGAGUAGAGCUCGGUCUCCCCAGUUAGGAGAGUGCUACCGCCGGCCGGAGUUCAAGCCC --------------.........((...((((......))))..(((((((((((((.((((.......))))(.(((((.......))))).))))))))))))))))........... ( -44.60, z-score = -2.19, R) >consensus AAUCAGACGAAAUUCAAGUUUAACCCAGGAUGCUGUCCCAUUGCCGCCGGCGGUAGUGGAGUGAGUAGAGCUCCGACACCCCAGUCAGUAGAGCACUACCGCCGGCCGGAGUUCAAGCCC ........(((.(((............((........))......(((((((((((((((((.......)))))(((......)))........)))))))))))).))).)))...... (-31.70 = -32.98 + 1.28)

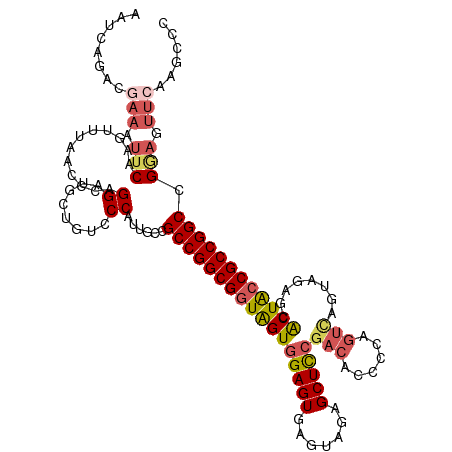
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:58:07 2011