| Sequence ID | dm3.chr2L |
|---|---|
| Location | 22,411,986 – 22,412,129 |
| Length | 143 |
| Max. P | 0.999762 |

| Location | 22,411,986 – 22,412,129 |
|---|---|
| Length | 143 |
| Sequences | 5 |
| Columns | 172 |
| Reading direction | forward |
| Mean pairwise identity | 72.16 |
| Shannon entropy | 0.50547 |
| G+C content | 0.53388 |
| Mean single sequence MFE | -56.20 |
| Consensus MFE | -33.70 |
| Energy contribution | -35.70 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.974106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
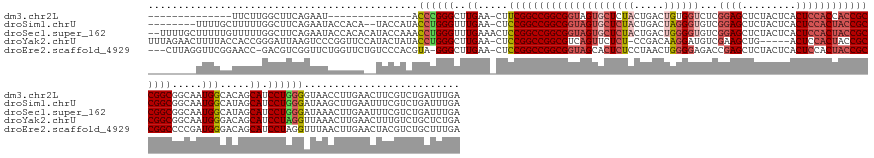
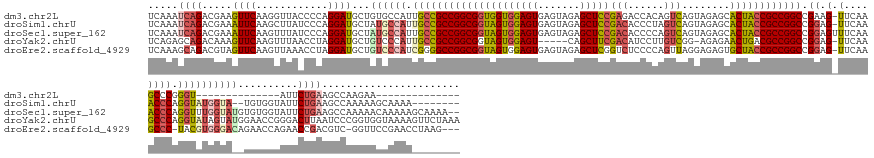
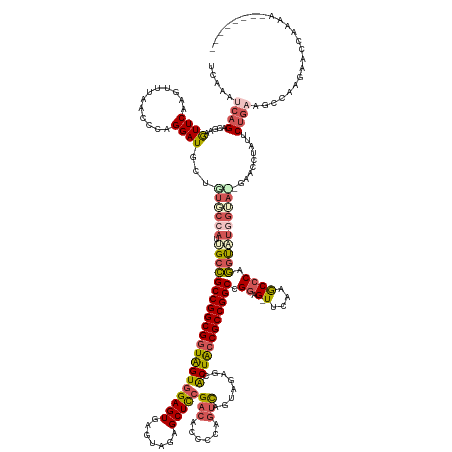
>dm3.chr2L 22411986 143 + 23011544 --------------UUCUUGGCUUCAGAAU--------------ACCCGGGCUUGAA-CUUCGGCCGGCGGUAGUGCUCUACUGACUGUGGUCUCGGAGCUCUACUCACUCCACCACCGCCGGCGGCAAUGGCACAGCAUCCUGGGGUAACCUUGAACUUCGUCUGAUUUGA --------------.........(((((..--------------.((((((..((..-.....(((((((((((.(((((...(((....)))..))))).))............))))))))).((....))....)).))))))((........))....)))))..... ( -46.50, z-score = -0.19, R) >droSim1.chrU 12456498 161 - 15797150 --------UUUUGCUUUUUGGCUUCAGAAUACCACA--UACCAUACCUGGGUUUGAA-CUCCGGCCGGCGGUAGUGCUCUACUGACUAGGGUGUCGGAGCUCUACUCACUCCACUACCGCCGGCGGCAAUGGCAUAGCAUCCUGGGAUAAGCUUGAAUUUCGUCUGAUUUGA --------....(((....))).(((((........--..((((.(((((((....)-).)))(((((((((((((......(((.((((((......)))))).)))...)))))))))))))))..))))((.((((((....)))..))))).......)))))..... ( -53.00, z-score = -1.60, R) >droSec1.super_162 22550 170 + 37850 --UUUUGCUUUUUGUUUUUGGCUUCAGAAUACCACACAUACCAAACCUGGGUUUGAAACUCCGGCCGGCGGUAGUGCUCUACUGACUGGGGUGUCGGAGCUCUACUCACUCCACUACCGCCGGCGGCAAUGGCAUAGCAUCCUGGGAUAAACUUGAAUUUCGUCUGAUUUGA --....(((...(((..(((.(((((((...(((.............))).))))))......(((((((((((((......(((.((((((......)))))).)))...)))))))))))))).)))..))).)))(((....)))....(..((((......))))..) ( -54.12, z-score = -1.72, R) >droYak2.chrU 23210840 165 - 28119190 UUUAGAACUUUUACCACCGGGAUUAAGUCCCGGUUCCAUACUAUACCUGGGCUUGAA-CUCCGGCCGGCGUCAGUUCUCU-CCGACAAGGAUGUCGAAGCUG-----ACUCCACUACCGCCGGCGGCAAUGGGACAGCAUCCUAGGUUAAACUUGAACUUUGUCUGCUCUGA .(((((.......(((.((((.((((((((.(((..........))).)))))))).-).)))(((((((..(((((.((-.(((((....))))).))..)-----)....)))..))))))).....)))....(((..(.(((((.......))))).)..)))))))) ( -53.00, z-score = -1.32, R) >droEre2.scaffold_4929 12731544 166 - 26641161 ---CUUAGGUUCGGAACC-GACGUCGGUUCUGGUUCUGUCCCACGUA-GGGCUUGAA-CUCCGGCCGGCGGUAGCACUCUCCUAACUGGGGAGACCGAGCUCUACUCACUCCACUACCGCCGGCCCCGAUGGGACAGCAUCCUAGGUUUAACUUGAACUACGUCUGCUUUGA ---((((((.((((((((-(....)))))))))..((((((((((..-(((......-.)))((((((((((((..((((((.....))))))...(((.....)))......)))))))))))).)).))))))))...)))))).......................... ( -74.40, z-score = -4.28, R) >consensus ________UUUUGCUUCUUGGCUUCAGAAUACCACA__UACCAUACCUGGGCUUGAA_CUCCGGCCGGCGGUAGUGCUCUACUGACUGGGGUGUCGGAGCUCUACUCACUCCACUACCGCCGGCGGCAAUGGCACAGCAUCCUGGGAUAAACUUGAACUUCGUCUGAUUUGA .............................................((((((..((.....(((((((((((((((((((((.....))))))...((((.........)))))))))))))))).....))).....)).)))))).......................... (-33.70 = -35.70 + 2.00)
| Location | 22,411,986 – 22,412,129 |
|---|---|
| Length | 143 |
| Sequences | 5 |
| Columns | 172 |
| Reading direction | reverse |
| Mean pairwise identity | 72.16 |
| Shannon entropy | 0.50547 |
| G+C content | 0.53388 |
| Mean single sequence MFE | -62.34 |
| Consensus MFE | -35.02 |
| Energy contribution | -37.22 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.30 |
| Mean z-score | -3.72 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.33 |
| SVM RNA-class probability | 0.999762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
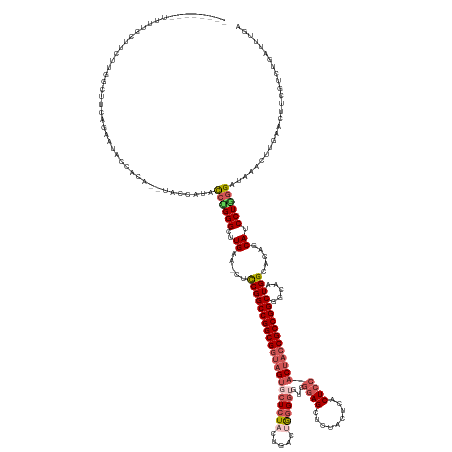
>dm3.chr2L 22411986 143 - 23011544 UCAAAUCAGACGAAGUUCAAGGUUACCCCAGGAUGCUGUGCCAUUGCCGCCGGCGGUGGUGGAGUGAGUAGAGCUCCGAGACCACAGUCAGUAGAGCACUACCGCCGGCCGAAG-UUCAAGCCCGGGU--------------AUUCUGAAGCCAAGAA-------------- .....(((((.(((.(((..(((....((((....))).).....)))(((((((((((((((((.......)))))..(((....)))........)))))))))))).))).-)))..((....))--------------..))))).........-------------- ( -51.00, z-score = -2.05, R) >droSim1.chrU 12456498 161 + 15797150 UCAAAUCAGACGAAAUUCAAGCUUAUCCCAGGAUGCUAUGCCAUUGCCGCCGGCGGUAGUGGAGUGAGUAGAGCUCCGACACCCUAGUCAGUAGAGCACUACCGCCGGCCGGAG-UUCAAACCCAGGUAUGGUA--UGUGGUAUUCUGAAGCCAAAAAGCAAAA-------- ....................((((....((((((((((((((((.((((((((((((((((((((.......)))))(((......)))........)))))))))))).((.(-(....)))).)))))))).--.)))))))))))))))............-------- ( -62.50, z-score = -4.83, R) >droSec1.super_162 22550 170 - 37850 UCAAAUCAGACGAAAUUCAAGUUUAUCCCAGGAUGCUAUGCCAUUGCCGCCGGCGGUAGUGGAGUGAGUAGAGCUCCGACACCCCAGUCAGUAGAGCACUACCGCCGGCCGGAGUUUCAAACCCAGGUUUGGUAUGUGUGGUAUUCUGAAGCCAAAAACAAAAAGCAAAA-- .......((((.........))))....(((((((((((((.....(((((((((((((((((((.......)))))(((......)))........))))))))))).)))....((((((....))))))...)))))))))))))......................-- ( -59.50, z-score = -3.86, R) >droYak2.chrU 23210840 165 + 28119190 UCAGAGCAGACAAAGUUCAAGUUUAACCUAGGAUGCUGUCCCAUUGCCGCCGGCGGUAGUGGAGU-----CAGCUUCGACAUCCUUGUCGG-AGAGAACUGACGCCGGCCGGAG-UUCAAGCCCAGGUAUAGUAUGGAACCGGGACUUAAUCCCGGUGGUAAAAGUUCUAAA ..(((((.((((..((((.((......)).))))..))))(((((((((....))))))))).((-----(((((((((((....))))))-))....)))))((((.((((..-((.((((((.(((..........)))))).)))))..))))))))....)))))... ( -64.60, z-score = -3.52, R) >droEre2.scaffold_4929 12731544 166 + 26641161 UCAAAGCAGACGUAGUUCAAGUUAAACCUAGGAUGCUGUCCCAUCGGGGCCGGCGGUAGUGGAGUGAGUAGAGCUCGGUCUCCCCAGUUAGGAGAGUGCUACCGCCGGCCGGAG-UUCAAGCCC-UACGUGGGACAGAACCAGAACCGACGUC-GGUUCCGAACCUAAG--- ..............((((............((...((((((((.((.(((((((((((((.((((.......))))(.(((((.......))))).))))))))))))))((.(-(....))))-..))))))))))..)).((((((....)-))))).)))).....--- ( -74.10, z-score = -4.35, R) >consensus UCAAAUCAGACGAAGUUCAAGUUUAACCCAGGAUGCUGUGCCAUUGCCGCCGGCGGUAGUGGAGUGAGUAGAGCUCCGACACCCCAGUCAGUAGAGCACUACCGCCGGCCGGAG_UUCAAGCCCAGGUAUGGUA__GAACCUAUUCUGAAGCCAAGAACCAAAA________ .....((((.....((((............))))...((((((.(((((((((((((((((((((.......)))))(((......)))........)))))))))))).((..........)).))))))))))..........))))....................... (-35.02 = -37.22 + 2.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:58:03 2011