| Sequence ID | dm3.chr2L |
|---|---|
| Location | 22,407,806 – 22,407,924 |
| Length | 118 |
| Max. P | 0.998595 |

| Location | 22,407,806 – 22,407,924 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 67.63 |
| Shannon entropy | 0.58348 |
| G+C content | 0.53394 |
| Mean single sequence MFE | -37.98 |
| Consensus MFE | -20.80 |
| Energy contribution | -23.16 |
| Covariance contribution | 2.36 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.955206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
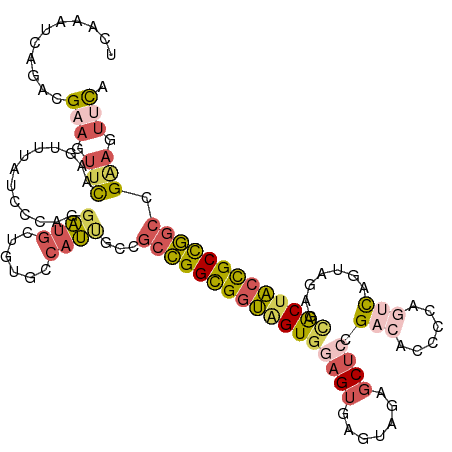
>dm3.chr2L 22407806 118 + 23011544 UGAACUUCGGCCGGCGGUAGUGCUCUACUGACUGGGGUCUCGCAGCUCUACUCACUCCACCACCGCCGGCGGCAAUGGCACAGCAUCCUGGGGUAACCUUGAACUUCGUCUGAUUUGA .(((.(((((((((((((.(((......(((.((((((......)))))).)))...))).)))))))))((.....((.(((....)))..))..)).)))).)))........... ( -37.60, z-score = 0.12, R) >droSim1.chr2h_random 780520 118 - 3178526 UGAACUUCGGCCGGCGGUAGUGCUCUACUGACUGGGGUGUCGGAGCUCUACUCACUCCACUACCGCCGGCGGCAAUGUCAUAGCAUCCUGGGAUAAACUUGAAUUUCGUCUGAUUUGA .(((.(((((((((((((((((......(((.((((((......)))))).)))...))))))))))))).....((((.(((....))).))))....)))).)))........... ( -42.30, z-score = -2.19, R) >droSec1.super_162 12836 118 + 37850 GAAACUCCGGCCGGCGGUAGUGCUCUACUGACUGGGGUGUCGGAGCUCUACUCACUCCACUACCGCCGGCGGCAAUGGCAUAGCAUCCUGGGAUAAACUUGAAUUUCGUCUGAUUUGA .....(((.(((((((((((((......(((.((((((......)))))).)))...))))))))))))).((....))...........))).....(..((((......))))..) ( -41.00, z-score = -1.45, R) >droEre2.scaffold_4929 12728927 118 - 26641161 UGAACUCCGGCCGGCGGUAGCACUCUCCUAACUGGGGAGACCGAGCUCUACUCACUCCACUACCGCCGGCCCCGAUGGGACAGCAUCCUAGGUUUAACUUGAACUACGUUUGCUUUGA .((((...((((((((((((..((((((.....))))))...(((.....)))......))))))))))))....(((((.....)))))(((((.....)))))..))))....... ( -46.30, z-score = -3.67, R) >droAna3.scaffold_13228 1071623 111 - 1099307 UGACCAGCUUAUAGCAGAUGUGGUCUAAUG-UUGAUAAUUCACCGUCUUUAUUGCUUAACUCGCACCAGUCGGGCAUGUUCGACAAGUCACUCGGCAAAUAAACCACUUUAG------ ......((.....))....(((((.((.((-((((.......(((.((....(((.......)))..)).)))......)))))).(((....)))...)).))))).....------ ( -22.72, z-score = -0.07, R) >consensus UGAACUCCGGCCGGCGGUAGUGCUCUACUGACUGGGGUGUCGGAGCUCUACUCACUCCACUACCGCCGGCGGCAAUGGCACAGCAUCCUGGGAUAAACUUGAACUUCGUCUGAUUUGA .........(((((((((((((......(((.((((((......)))))).)))...)))))))))))))................................................ (-20.80 = -23.16 + 2.36)


| Location | 22,407,806 – 22,407,924 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 67.63 |
| Shannon entropy | 0.58348 |
| G+C content | 0.53394 |
| Mean single sequence MFE | -40.33 |
| Consensus MFE | -23.06 |
| Energy contribution | -24.66 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.998595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 22407806 118 - 23011544 UCAAAUCAGACGAAGUUCAAGGUUACCCCAGGAUGCUGUGCCAUUGCCGCCGGCGGUGGUGGAGUGAGUAGAGCUGCGAGACCCCAGUCAGUAGAGCACUACCGCCGGCCGAAGUUCA ...........(((.(((..(((....((((....))).).....)))(((((((((((((............((((..(((....))).))))..))))))))))))).))).))). ( -42.74, z-score = -1.91, R) >droSim1.chr2h_random 780520 118 + 3178526 UCAAAUCAGACGAAAUUCAAGUUUAUCCCAGGAUGCUAUGACAUUGCCGCCGGCGGUAGUGGAGUGAGUAGAGCUCCGACACCCCAGUCAGUAGAGCACUACCGCCGGCCGAAGUUCA ...........(((.(((..(((((((....))))....)))......(((((((((((((((((.......)))))(((......)))........)))))))))))).))).))). ( -42.80, z-score = -3.96, R) >droSec1.super_162 12836 118 - 37850 UCAAAUCAGACGAAAUUCAAGUUUAUCCCAGGAUGCUAUGCCAUUGCCGCCGGCGGUAGUGGAGUGAGUAGAGCUCCGACACCCCAGUCAGUAGAGCACUACCGCCGGCCGGAGUUUC ...........(((((((.(((..(((....))))))...........(((((((((((((((((.......)))))(((......)))........))))))))))))..))))))) ( -44.00, z-score = -3.78, R) >droEre2.scaffold_4929 12728927 118 + 26641161 UCAAAGCAAACGUAGUUCAAGUUAAACCUAGGAUGCUGUCCCAUCGGGGCCGGCGGUAGUGGAGUGAGUAGAGCUCGGUCUCCCCAGUUAGGAGAGUGCUACCGCCGGCCGGAGUUCA ....((((..(...(((.......)))...)..)))).(((......(((((((((((((.((((.......))))(.(((((.......))))).)))))))))))))))))..... ( -44.80, z-score = -1.98, R) >droAna3.scaffold_13228 1071623 111 + 1099307 ------CUAAAGUGGUUUAUUUGCCGAGUGACUUGUCGAACAUGCCCGACUGGUGCGAGUUAAGCAAUAAAGACGGUGAAUUAUCAA-CAUUAGACCACAUCUGCUAUAAGCUGGUCA ------.....((((((((((..(((......((((..(((.(((((....)).))).)))..))))......)))..)).......-...))))))))(((.((.....)).))).. ( -27.30, z-score = -0.60, R) >consensus UCAAAUCAGACGAAGUUCAAGUUUAUCCCAGGAUGCUGUGCCAUUGCCGCCGGCGGUAGUGGAGUGAGUAGAGCUCCGACACCCCAGUCAGUAGAGCACUACCGCCGGCCGAAGUUCA ...........(((.(((.............((((......))))...(((((((((((((((((.......)))).(((......))).......))))))))))))).))).))). (-23.06 = -24.66 + 1.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:58:01 2011