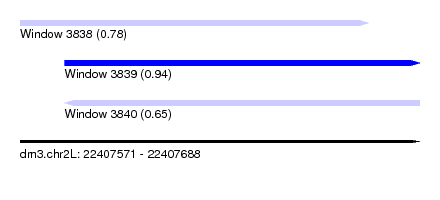
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 22,407,571 – 22,407,688 |
| Length | 117 |
| Max. P | 0.942748 |

| Location | 22,407,571 – 22,407,673 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 78.56 |
| Shannon entropy | 0.37196 |
| G+C content | 0.41668 |
| Mean single sequence MFE | -23.12 |
| Consensus MFE | -14.70 |
| Energy contribution | -14.78 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.779063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 22407571 102 + 23011544 -AAAAUCAUAAAUUUGGCGGGACGAACGCACAAACUUU-CUGAUCUAGCCGC-UGCAAUCCGUAGCGAGCUAAAUAA-GAAAAUCAGUUCGUUACAGAUUGAUAAA -...((((...(((((....(((((((........(((-((.((.(((((((-(((.....)))))).)))).)).)-))))....))))))).)))))))))... ( -29.90, z-score = -3.62, R) >droSec1.super_69 23438 93 + 174750 -CAAAUCAUAAAUUUGGUGGGACGAACGCACAAACUCU-CUGAGCUAGCCGC-UGCAACCAGUAGCGAGCGAAAUAAAGAAAAUCAGUUCGUUACA---------- -...((((......))))(..((((((((.((......-.)).))..(((((-(((.....)))))).))................))))))..).---------- ( -24.50, z-score = -2.08, R) >droYak2.chr2h_random 792673 105 - 3774259 UUAUUCUAUAAAUUUGGUGGGACGAACGCACAAACUCUACUGAGCUAGCCGCACGUAUUCCGUAGCGAGC-AGACAAACAAAAUCAGUUCGUUACAAGAAUAUGAA .((((((..........((..((((((.......(((....)))...((((((((.....))).))).))-...............))))))..)))))))).... ( -23.30, z-score = -1.20, R) >droEre2.scaffold_4784 23980780 90 + 25762168 -CAAAUCAUAAAUUUGGUGGAACGAACAAACAAACUCU-CUGAGCUAGCCGU-UGAAAUUCGUAGCGAGCAGAACCAAGAAAAACAGUUCGUU------------- -...((((......))))..(((((((..........(-(((.((((..((.-.......))))))...)))).....(.....).)))))))------------- ( -15.10, z-score = 0.08, R) >droSim1.chrU 8921692 93 - 15797150 -CAAAUCAUAAAUUUGGUGGGACGAACACACAAACUCU-CUGAGCUAGCCGC-UGCAACCAGUAGCGAGCGAAAUAAAGAAAAUCGGUUCGUUACA---------- -...((((......))))(..((((((.......(((.-..)))...(((((-(((.....)))))).))................))))))..).---------- ( -22.80, z-score = -1.69, R) >consensus _CAAAUCAUAAAUUUGGUGGGACGAACGCACAAACUCU_CUGAGCUAGCCGC_UGCAAUCCGUAGCGAGCGAAAUAAAGAAAAUCAGUUCGUUACA__________ ....((((......))))..(((((((.......(((....)))...(((((.(((.....)))))).))................)))))))............. (-14.70 = -14.78 + 0.08)
| Location | 22,407,584 – 22,407,688 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 72.18 |
| Shannon entropy | 0.44260 |
| G+C content | 0.44844 |
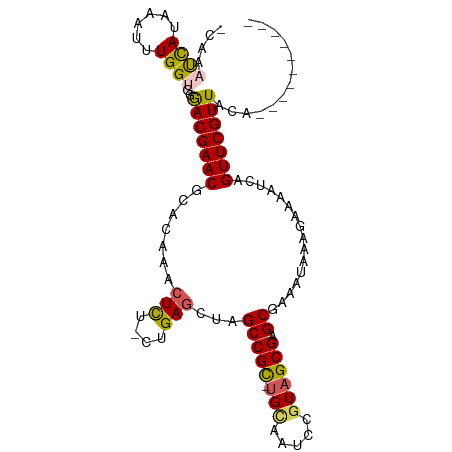
| Mean single sequence MFE | -24.05 |
| Consensus MFE | -15.98 |
| Energy contribution | -16.10 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.942748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
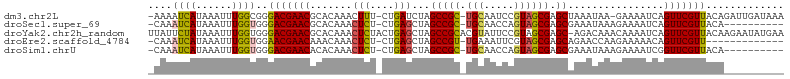
>dm3.chr2L 22407584 104 + 23011544 UGGCGGGACGAACGCACAAACUUU-CUGAUCUAGCCGC-UGCAAUCCGUAGCGAGCUAAAUAA-GAAAAUCAGUUCGUUACAGAUUGAUAAAAUAAAAUGUGUAUAU ......(((((((........(((-((.((.(((((((-(((.....)))))).)))).)).)-))))....)))))))(((.(((..........))).))).... ( -27.30, z-score = -2.38, R) >droSec1.super_69 23451 80 + 174750 UGGUGGGACGAACGCACAAACUCU-CUGAGCUAGCCGC-UGCAACCAGUAGCGAGCGAAAUAAAGAAAAUCAGUUCGUUACA------------------------- ...((..((((((((.((......-.)).))..(((((-(((.....)))))).))................))))))..))------------------------- ( -23.40, z-score = -1.66, R) >droYak2.chr2h_random 792687 106 - 3774259 UGGUGGGACGAACGCACAAACUCUACUGAGCUAGCCGCACGUAUUCCGUAGCGAGC-AGACAAACAAAAUCAGUUCGUUACAAGAAUAUGAAGCAAAGAAAUAAAAU ..(((.(.....).)))...(((....)))......((.(((((((.(((((((((-.((.........)).)))))))))..)))))))..))............. ( -23.80, z-score = -1.58, R) >droSim1.chrU 8921705 80 - 15797150 UGGUGGGACGAACACACAAACUCU-CUGAGCUAGCCGC-UGCAACCAGUAGCGAGCGAAAUAAAGAAAAUCGGUUCGUUACA------------------------- ...((..((((((.......(((.-..)))...(((((-(((.....)))))).))................))))))..))------------------------- ( -21.70, z-score = -1.28, R) >consensus UGGUGGGACGAACGCACAAACUCU_CUGAGCUAGCCGC_UGCAACCAGUAGCGAGCGAAAUAAAGAAAAUCAGUUCGUUACA_________________________ ....(..((((((.......(((....)))...(((((.(((.....)))))).))................))))))..).......................... (-15.98 = -16.10 + 0.12)

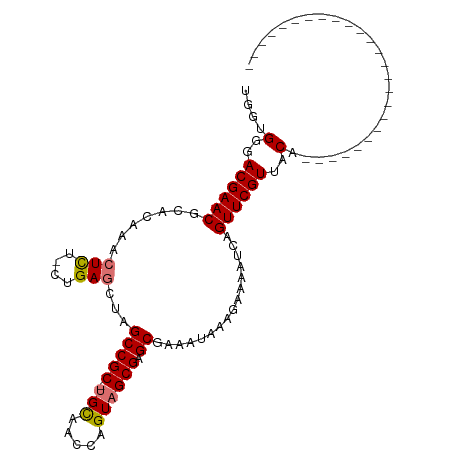
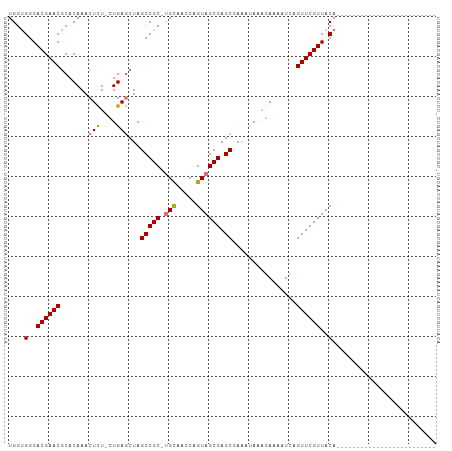
| Location | 22,407,584 – 22,407,688 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 72.18 |
| Shannon entropy | 0.44260 |
| G+C content | 0.44844 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -14.52 |
| Energy contribution | -14.40 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.651363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 22407584 104 - 23011544 AUAUACACAUUUUAUUUUAUCAAUCUGUAACGAACUGAUUUUC-UUAUUUAGCUCGCUACGGAUUGCA-GCGGCUAGAUCAG-AAAGUUUGUGCGUUCGUCCCGCCA ..........................(..((((((.(((((((-(.(((((((.((((.(.....).)-)))))))))).))-)))))).....))))))..).... ( -26.90, z-score = -2.50, R) >droSec1.super_69 23451 80 - 174750 -------------------------UGUAACGAACUGAUUUUCUUUAUUUCGCUCGCUACUGGUUGCA-GCGGCUAGCUCAG-AGAGUUUGUGCGUUCGUCCCACCA -------------------------((..((((((.((((((((.......(((.((..(((....))-)..)).)))..))-)))))).....))))))..))... ( -21.30, z-score = -0.66, R) >droYak2.chr2h_random 792687 106 + 3774259 AUUUUAUUUCUUUGCUUCAUAUUCUUGUAACGAACUGAUUUUGUUUGUCU-GCUCGCUACGGAAUACGUGCGGCUAGCUCAGUAGAGUUUGUGCGUUCGUCCCACCA .........................((..((((((.(((.......))).-((.(((.(((.....))))))((.(((((....))))).))))))))))..))... ( -22.90, z-score = -0.47, R) >droSim1.chrU 8921705 80 + 15797150 -------------------------UGUAACGAACCGAUUUUCUUUAUUUCGCUCGCUACUGGUUGCA-GCGGCUAGCUCAG-AGAGUUUGUGUGUUCGUCCCACCA -------------------------((..((((((((...((((((.....(((.((..(((....))-)..)).)))..))-))))....)).))))))..))... ( -22.40, z-score = -1.36, R) >consensus _________________________UGUAACGAACUGAUUUUCUUUAUUUCGCUCGCUACGGAUUGCA_GCGGCUAGCUCAG_AGAGUUUGUGCGUUCGUCCCACCA .........................((..((((((................((.(((............)))))..((.(((......))).))))))))..))... (-14.52 = -14.40 + -0.12)

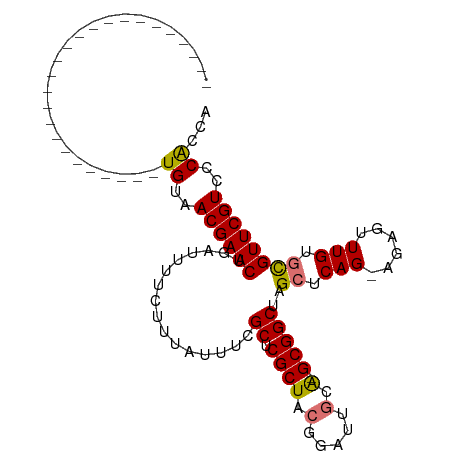
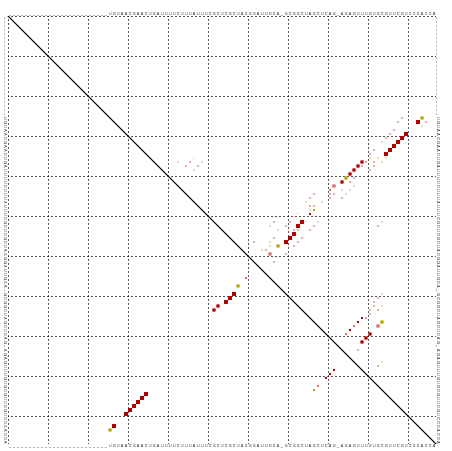
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:58:00 2011