
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 22,393,473 – 22,393,658 |
| Length | 185 |
| Max. P | 0.757072 |

| Location | 22,393,473 – 22,393,658 |
|---|---|
| Length | 185 |
| Sequences | 4 |
| Columns | 192 |
| Reading direction | reverse |
| Mean pairwise identity | 54.27 |
| Shannon entropy | 0.72704 |
| G+C content | 0.52344 |
| Mean single sequence MFE | -61.23 |
| Consensus MFE | -23.54 |
| Energy contribution | -25.23 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.757072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 22393473 185 - 23011544 ------AAGGAGGGAGUGCGACCUGGCCUCACAUCCUGCU-CACCGAAGUCAUACCUUGACUGGCAGUCCCGGUGAGCGAGCAAGGACUGAUGAACACGCGGAUGUUUUGGUUUUAGUACGUAGGCAUAAUUCCAAUAGGGCUUAUGAAUCGUGCAUGUCACCUACGGGCGGUAUGUGGUAUCUUUAGAAGG ------....(((((.(((.((..((((..((((((((((-(((((.(((((.....)))))((....))))))))))).((..(..(....)..)..))))))))...))))...(((((.(..(((((..((....))..)))))..))))))((..(.((....)).)..)))).))))))))...... ( -58.50, z-score = 0.20, R) >droAna3.scaffold_13230 258765 171 + 3602488 ACGUCGCUUAAGGGAGAAUGCCUAGGUUAGGCAACCCUUCCCCGCGAAGUAAUACCUUA-----------CAGUGGU-UCGCGGGGUCUGUGGUGUGUGC---GGGGGUGGUUUUAGUUCGUAGGCACUGGC-UAGACACGCUGGUGAAUCGUGCAUACUAUUGGCGCGUACCAA-UCUCUUCGUACG---- ((..(((..(((((....((((((...)))))).)))))((((((((......(((...-----------....)))-))))))))...)))..))((((---(((((((..((((((..((....))..))-))))..)))(((((...(((((.........)))))))))).-...)))))))).---- ( -63.50, z-score = -0.87, R) >droVir3.scaffold_12799 195909 168 + 516347 ------ACUAAUGGAUACUGCC-AAAAUAGGCAACCCAUCACCGCGAAUUAAUACUUUG-----------CGGUGGUCCCGCGGGGAAUGUGGUGUGUGAGUGGGGGGUGAGUUUAGUACGUAGGUGCUGAC-----GGAGUCAGCAAAUCGUGCAUAUGGUUGGCCCGGGCCAU-CAGUAUCUUACGAAGA ------......((((((((((-(...(((((.(((((((((((((((.......))))-----------)))))))(((((.....((.....))....)))))))))..)))))(((((....(((((((-----...)))))))...)))))...))).((((....)))).-))))))))........ ( -65.40, z-score = -2.48, R) >droGri2.scaffold_15203 933029 162 - 11997470 ------AGUGACGGAGAGUGCC-----UGGGCAACUUAUC-CCG-GAAGUAACACUUUA-----------CGGUGAUACCGCGGGGGAUUGGGGAUGUGUGUGGGGGAUGAGUUUAGUACGAAUGCAUUGAU-----GUUGUCUUUGAAGCGUGCAUGUAGUUAGCCCGGGCCUU-CUGUAUCUCCAGGAGA ------....(((((.((.(((-----(((((((((((((-((.-.......((((...-----------.))))...(((((.(.(........).).)))))))))))))))..(((((....((..(((-----...)))..))...))))).........)))))))))))-)))).((((...)))) ( -57.50, z-score = -1.51, R) >consensus ______AAUAAGGGAGAGUGCC_AGG_UAGGCAACCCAUC_CCGCGAAGUAAUACCUUA___________CGGUGAUCCCGCGGGGAAUGUGGUAUGUGCGUGGGGGGUGAGUUUAGUACGUAGGCACUGAC_____GGAGCCUGUGAAUCGUGCAUAUAAUUGGCCCGGGCCAU_CAGUAUCUUAAGAAGA .............(((((((.......(((((((((((((.((.........((((...............))))...(((((.....)))))..........)).))))))))..(((((....(((((((........)))))))...))))).........))))).......)))))))......... (-23.54 = -25.23 + 1.69)

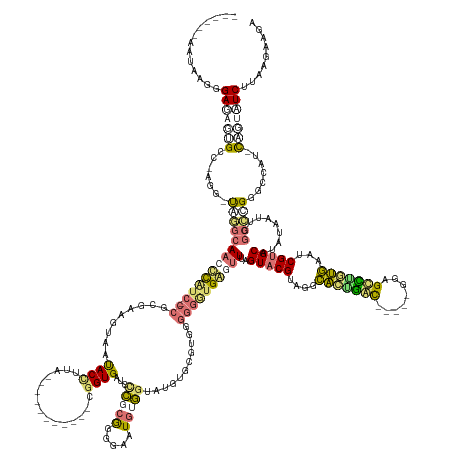
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:57:57 2011