
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 22,269,834 – 22,269,944 |
| Length | 110 |
| Max. P | 0.999164 |

| Location | 22,269,834 – 22,269,944 |
|---|---|
| Length | 110 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 68.13 |
| Shannon entropy | 0.61558 |
| G+C content | 0.50995 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -15.09 |
| Energy contribution | -16.20 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.990190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
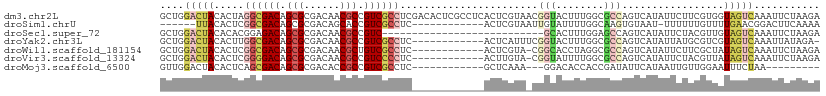
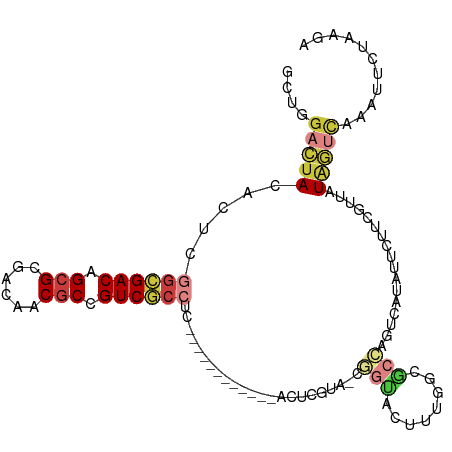
>dm3.chr2L 22269834 110 + 23011544 GCUGGACUACACUAGGCGACAGCGCGACAACGCCGUCGCCUCGACACUCGCCUCACUCGUAACGGUACUUUGGCGCCAGUCAUAUUCUUCGUGGUAGUCAAAUUCUAAGA ....((((((((.(((((((.(((......))).))))))).(((...((((..(((......))).....))))...))).........)).))))))........... ( -35.00, z-score = -1.65, R) >droSim1.chrU 10082218 91 - 15797150 ------UUACACUCGGCGACAGCGCGACAGCACCGUCGCCUC------------ACUCGUAAUUGUAUUUUGGCAAGUGUAAU-UUUUUUGUUUUGAACGGACUUCAAAA ------((((((..((((((.(.((....)).).))))))..------------........((((......)))))))))).-.......(((((((.....))))))) ( -24.50, z-score = -1.60, R) >droSec1.super_72 42855 83 - 149286 GCUGGACUACACACGGAGACAGCGCGACAACGCCGUC---------------------------GCACUUUGGAGCCAGUCAUAUUCUACGUUGUAGUCAAAUUCUAAGA ....(((((((.(((.(((.((.(((((......)))---------------------------)).))(((....)))......))).))))))))))........... ( -23.90, z-score = -1.54, R) >droYak2.chr3L 23750628 97 + 24197627 GCUGGACUACACUUGGCGACAGCGCGACAACGCCGUCGCCUC------------ACUCAUUUCGGUACUUUGGCGCCAGUCAUAUUAUGCGUCGUAGUCAAAUUAUAGA- ....((((((....((((((.(((......))).))))))..------------.................(((((............)))))))))))..........- ( -30.90, z-score = -1.89, R) >droWil1.scaffold_181154 2779707 97 - 4610121 GCUGGACUACACUCGGCGACAGCGCGACAACGCUGUCGCCUC------------ACUCGUA-CGGCACCUAGGCGCCAGUCAUAUUCUUCGCUAUAGUCAAAUUCUAAGA ....(((((.....((((((((((......))))))))))..------------.......-.(((.(....).))).................)))))........... ( -32.70, z-score = -2.77, R) >droVir3.scaffold_13324 1039760 97 + 2960039 GCUGGACUACACUCGGGGACAGCGCGACAACGCCGUCCCCUC------------ACUUGUA-CGGUAUUUUGGCGCCAGUCAUAUUCUACGUUAUAGUCAAAUUCUAAGA ....(((((.....((((((.(((......))).))))))..------------....(((-..(((...((((....)))))))..)))....)))))........... ( -28.20, z-score = -1.76, R) >droMoj3.scaffold_6500 1112820 86 - 32352404 GUUGGACUACACUCAGCGACAGCGCGACACCGCCGUCGCCUC------------GCUCAAA---GGACACCACCGAUAUUCAUAAUUGUUGGAAUUUCUAA--------- (.((..((......(((((..(.(((((......))))))))------------)))...)---)..)).).((((((........)))))).........--------- ( -20.60, z-score = -1.16, R) >consensus GCUGGACUACACUCGGCGACAGCGCGACAACGCCGUCGCCUC____________ACUCGUA_CGGUACUUUGGCGCCAGUCAUAUUCUUCGUUAUAGUCAAAUUCUAAGA ....(((((.....((((((.(((......))).)))))).......................(((........))).................)))))........... (-15.09 = -16.20 + 1.11)
| Location | 22,269,834 – 22,269,944 |
|---|---|
| Length | 110 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 68.13 |
| Shannon entropy | 0.61558 |
| G+C content | 0.50995 |
| Mean single sequence MFE | -33.19 |
| Consensus MFE | -21.24 |
| Energy contribution | -21.86 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.68 |
| SVM RNA-class probability | 0.999164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
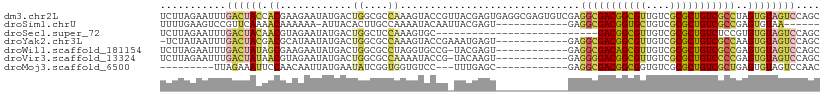
>dm3.chr2L 22269834 110 - 23011544 UCUUAGAAUUUGACUACCACGAAGAAUAUGACUGGCGCCAAAGUACCGUUACGAGUGAGGCGAGUGUCGAGGCGACGGCGUUGUCGCGCUGUCGCCUAGUGUAGUCCAGC ...........((((((.((........((((...((((....(((((...)).))).))))...))))((((((((((((....)))))))))))).)))))))).... ( -43.50, z-score = -2.79, R) >droSim1.chrU 10082218 91 + 15797150 UUUUGAAGUCCGUUCAAAACAAAAAA-AUUACACUUGCCAAAAUACAAUUACGAGU------------GAGGCGACGGUGCUGUCGCGCUGUCGCCGAGUGUAA------ (((((((.....))))))).......-.((((((((.......(((........))------------).(((((((((((....)))))))))))))))))))------ ( -32.40, z-score = -3.95, R) >droSec1.super_72 42855 83 + 149286 UCUUAGAAUUUGACUACAACGUAGAAUAUGACUGGCUCCAAAGUGC---------------------------GACGGCGUUGUCGCGCUGUCUCCGUGUGUAGUCCAGC ...........((((((((((.(((.......((....)).(((((---------------------------((((....))))))))).))).))).))))))).... ( -26.60, z-score = -1.76, R) >droYak2.chr3L 23750628 97 - 24197627 -UCUAUAAUUUGACUACGACGCAUAAUAUGACUGGCGCCAAAGUACCGAAAUGAGU------------GAGGCGACGGCGUUGUCGCGCUGUCGCCAAGUGUAGUCCAGC -..........(((((((((.(((......(((........)))......))).))------------..(((((((((((....)))))))))))...))))))).... ( -33.40, z-score = -1.97, R) >droWil1.scaffold_181154 2779707 97 + 4610121 UCUUAGAAUUUGACUAUAGCGAAGAAUAUGACUGGCGCCUAGGUGCCG-UACGAGU------------GAGGCGACAGCGUUGUCGCGCUGUCGCCGAGUGUAGUCCAGC ...........(((((((...............(((((....))))).-.......------------..(((((((((((....)))))))))))...))))))).... ( -39.30, z-score = -2.95, R) >droVir3.scaffold_13324 1039760 97 - 2960039 UCUUAGAAUUUGACUAUAACGUAGAAUAUGACUGGCGCCAAAAUACCG-UACAAGU------------GAGGGGACGGCGUUGUCGCGCUGUCCCCGAGUGUAGUCCAGC ...........(((((((..........(.(((.(((.........))-)...)))------------.)(((((((((((....)))))))))))...))))))).... ( -32.40, z-score = -2.25, R) >droMoj3.scaffold_6500 1112820 86 + 32352404 ---------UUAGAAAUUCCAACAAUUAUGAAUAUCGGUGGUGUCC---UUUGAGC------------GAGGCGACGGCGGUGUCGCGCUGUCGCUGAGUGUAGUCCAAC ---------...((.((((..........)))).)).....((..(---(....((------------..((((((((((......))))))))))..))..))..)).. ( -24.70, z-score = -0.40, R) >consensus UCUUAGAAUUUGACUACAACGAAGAAUAUGACUGGCGCCAAAGUACCG_UACGAGU____________GAGGCGACGGCGUUGUCGCGCUGUCGCCGAGUGUAGUCCAGC ...........((((((.((............((....))..............................(((((((((((....)))))))))))..)))))))).... (-21.24 = -21.86 + 0.62)
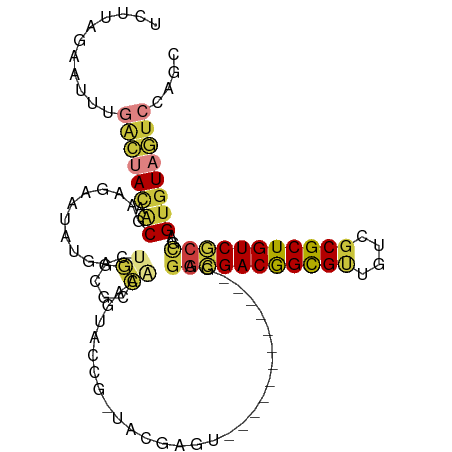
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:57:46 2011