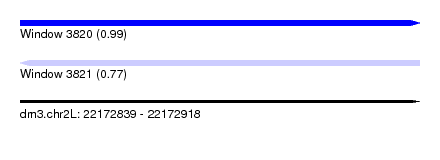
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 22,172,839 – 22,172,918 |
| Length | 79 |
| Max. P | 0.986426 |

| Location | 22,172,839 – 22,172,918 |
|---|---|
| Length | 79 |
| Sequences | 3 |
| Columns | 82 |
| Reading direction | forward |
| Mean pairwise identity | 59.92 |
| Shannon entropy | 0.53953 |
| G+C content | 0.50602 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -12.38 |
| Energy contribution | -14.83 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.986426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
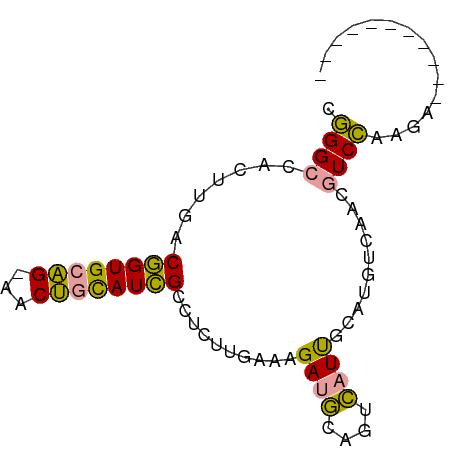
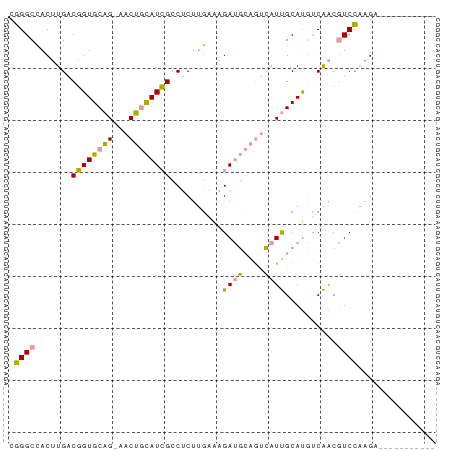
>dm3.chr2L 22172839 79 + 23011544 CAGGCUACGUG-CUGUGCUG-GACAAUAUGGACACUGAAGUGAAUGAAAAAUCGCAC-UCGGCUUCUCCGACAUGCAUAAAU ........(((-(.(((.((-((.......((..((((.((((.(....).))))..-))))..)))))).))))))).... ( -15.01, z-score = 0.43, R) >dp4.Unknown_group_143 3347 70 + 20963 CGGGCCACUUGACGGUGCAGAAACUGCAUCGCCUCUUGAAAGAUGCAGUCAUUGCAUGCCAACGUCCAAG------------ .((((...(((.((.(((((..((((((((...........))))))))..))))))).))).))))...------------ ( -24.40, z-score = -2.28, R) >droPer1.super_62 70662 73 - 349094 CGGGCCACUUGACGGUGCAGUAACUGCAUCGCCUCUUGAAAGAUGCAGUCAUUGCAUGUCAACGUCCAAGAAU--------- .((((...((((((.((((((.((((((((...........)))))))).)))))))))))).))))......--------- ( -31.40, z-score = -4.56, R) >consensus CGGGCCACUUGACGGUGCAG_AACUGCAUCGCCUCUUGAAAGAUGCAGUCAUUGCAUGUCAACGUCCAAGA___________ .((((...(((...((((((..((((((((...........))))))))..))))))..))).))))............... (-12.38 = -14.83 + 2.45)

| Location | 22,172,839 – 22,172,918 |
|---|---|
| Length | 79 |
| Sequences | 3 |
| Columns | 82 |
| Reading direction | reverse |
| Mean pairwise identity | 59.92 |
| Shannon entropy | 0.53953 |
| G+C content | 0.50602 |
| Mean single sequence MFE | -22.86 |
| Consensus MFE | -9.97 |
| Energy contribution | -8.87 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.774559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
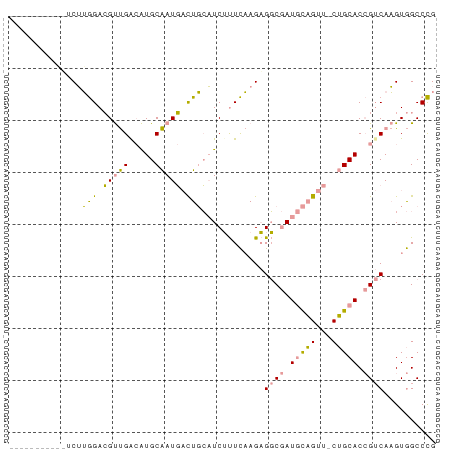
>dm3.chr2L 22172839 79 - 23011544 AUUUAUGCAUGUCGGAGAAGCCGA-GUGCGAUUUUUCAUUCACUUCAGUGUCCAUAUUGUC-CAGCACAG-CACGUAGCCUG .....(((...((((.....))))-(((((((........(((....)))........)))-..)))).)-))......... ( -16.09, z-score = -0.09, R) >dp4.Unknown_group_143 3347 70 - 20963 ------------CUUGGACGUUGGCAUGCAAUGACUGCAUCUUUCAAGAGGCGAUGCAGUUUCUGCACCGUCAAGUGGCCCG ------------...((.(((((((.((((..((((((((((((....))).)))))))))..))))..))))).))..)). ( -25.40, z-score = -1.86, R) >droPer1.super_62 70662 73 + 349094 ---------AUUCUUGGACGUUGACAUGCAAUGACUGCAUCUUUCAAGAGGCGAUGCAGUUACUGCACCGUCAAGUGGCCCG ---------......((.(((((((.((((.(((((((((((((....))).)))))))))).))))..))))).))..)). ( -27.10, z-score = -3.02, R) >consensus ___________UCUUGGACGUUGACAUGCAAUGACUGCAUCUUUCAAGAGGCGAUGCAGUU_CUGCACCGUCAAGUGGCCCG ..............(((.(((((.....))))).)))............((((.(((((...))))).)))).......... ( -9.97 = -8.87 + -1.10)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:57:44 2011