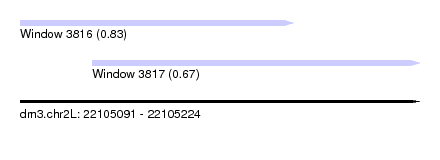
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 22,105,091 – 22,105,224 |
| Length | 133 |
| Max. P | 0.828957 |

| Location | 22,105,091 – 22,105,182 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 66.25 |
| Shannon entropy | 0.73216 |
| G+C content | 0.43769 |
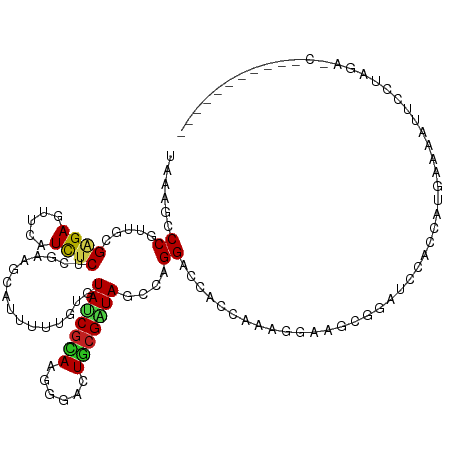
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -8.96 |
| Energy contribution | -8.70 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.828957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
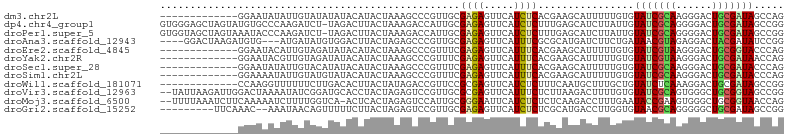
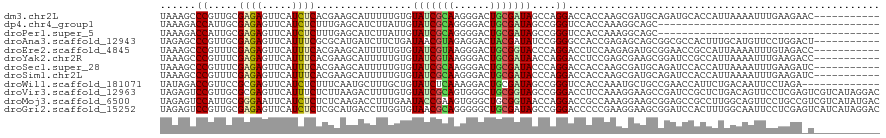
>dm3.chr2L 22105091 91 + 23011544 -------------GGAAUAUAUUGUAUAUAUACAUACUAAAGCCCGUUGCGAGAGUUCAUCUCACGAAGCAUUUUUGUGUAUCGCAAGGGACUGCGAUAGCCAG -------------((....((((((.(((....)))....(((((.((((((((.....))))..((.((((....)))).))))))))).)))))))).)).. ( -23.20, z-score = -1.63, R) >dp4.chr4_group1 1497727 103 + 5278887 GUGGGAGCUAGUAUGUGCCCAAGAUCU-UAGACUUACUAAAGACCAUUGCGAGAGUUCAUCUCUUUGAGCAUCUUAUUGUAUCGCAGGGGACUGCGAUAGCCGG .((((.((......)).)))).(.(((-(((.....)).)))).)...(((((((((((......))))).))))....((((((((....))))))))))... ( -33.00, z-score = -1.70, R) >droPer1.super_5 3103815 103 - 6813705 GUGGUAGCUAGUAAAUACCCAAGAUCU-UAGACUUACUAAAGACCAUUGCGAGAGUUCAUCUCUUUGAGCAUCUUAUUGUAUCGCAGGGGACUGCGAUAGCCGG (((((...((((((.............-.....))))))...))))).(((((((((((......))))).))))....((((((((....))))))))))... ( -31.97, z-score = -2.39, R) >droAna3.scaffold_12943 2297853 97 - 5039921 ----GGACUAAGAUGUG---AUGAUAUGUGGACUUACUAGAGCCCGUUGCGAGAGUUCAUUUCGCGCAUGAUCUUCUGAUAACGUAGAGGACUACGAUAUCCGG ----(((...(((...(---((..(((((((.(((....))).))...((((((.....)))))))))))))).))).....(((((....)))))...))).. ( -23.70, z-score = -0.20, R) >droEre2.scaffold_4845 2348531 91 + 22589142 -------------GGAAUACAUUGUAGAUAUACAUACUAAAGCCCGUUUCGAGAGUUCAUUUCACGAAGCAUUUUUGUGUAUCGUAAGGGACUGCGGUACCCAG -------------((.......((((....))))((((..(((((.....((((.....))))((((.((((....)))).))))..))).))..)))).)).. ( -21.80, z-score = -1.56, R) >droYak2.chr2R 650474 91 + 21139217 -------------GGAAUACGUUGUAGAUAUACAUACUAAAGCCCGUUUCGAGAGUUCAUUUCACGAAGCAUUUUUGUGUAUCGUAAGGGACUGCGAUAACCAG -------------((.((((((((((....)))).))........((((((.(((.....))).))))))......))))((((((......))))))..)).. ( -19.90, z-score = -0.98, R) >droSec1.super_28 709389 91 + 873875 -------------GGAAUAUAUUGUACAUAUACAUACUAAAGCCCGUUUCGAGAGUUCAUUUCACGAAGCAUUUUUGUGUAUCGCAAGGGACUGCGAUACCCAG -------------((.......((((....))))........)).((((((.(((.....))).)))))).....((.((((((((......)))))))).)). ( -22.86, z-score = -2.54, R) >droSim1.chr2L 21675728 91 + 22036055 -------------GGAAAAUAUUGUAUGUAUACAUACUAAAGCCCGUUUCGAGAGUUCAUUUCACGAAGCAUUUUUGUGUAUCGCAAGGGACUGCGAUACCCAG -------------.((((((...(((((....)))))........((((((.(((.....))).))))))))))))(.((((((((......)))))))).).. ( -25.90, z-score = -3.22, R) >droWil1.scaffold_181071 548258 91 + 1211509 -------------CCAAGGUUUUUUCUUGACACUUACUAUAGACCGUUCCGCGAGUUCAUCUCUUUCAAUGCUUUGCUGUAUCUCAAAGGACUGCGAUAGCCGG -------------.(((((.....)))))..............(((......(((.....))).......(((((((.((.((.....)))).)))).)))))) ( -14.30, z-score = 1.09, R) >droVir3.scaffold_12963 15043505 102 - 20206255 --UAUUAAGAUUGGACUAAAAUAUCGGAUGCACCUACUAGAGUCCGUUGCGCGAGUUCAUUUCUCUUAAGACUUUUGUGUAUCGCAGUGGGCUGCGGUAGCCGG --..((((((.((((((.......(((((.(........).))))).......))))))....))))))......((..((((((((....))))))))..)). ( -27.84, z-score = -0.66, R) >droMoj3.scaffold_6500 17431088 101 + 32352404 --UUUUAAAUCUUCAAAAAUCUUUUGGUCA-ACUCACUAGAGUCCAUUGCGGGAAUUCAUCUCUCUCAAGACCUUUGAAUACCGAAGUGGGCUGCGGUAACCAG --......................((((..-....(((..((((((((.(((..(((((....((....))....))))).))).))))))))..))).)))). ( -22.50, z-score = -1.00, R) >droGri2.scaffold_15252 9368100 93 - 17193109 ---------UUCAAAC--AAAUAACAGUUUUUCUUACUAGAGUCCGUUGCGAGAGUUCAUCUCUCGCAUGACCUUGGUGUAACGCAGGGGGCUGCGAUAGCCGG ---------.......--................((((((.(((...((((((((.....)))))))).))).))))))...(((((....)))))........ ( -28.20, z-score = -1.71, R) >consensus _____________GGAAUAUAUUGUAUAUAUACUUACUAAAGCCCGUUGCGAGAGUUCAUCUCUCGAAGCAUUUUUGUGUAUCGCAAGGGACUGCGAUAGCCAG ..................................................((((.....))))................(((((((......)))))))..... ( -8.96 = -8.70 + -0.26)
| Location | 22,105,115 – 22,105,224 |
|---|---|
| Length | 109 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.95 |
| Shannon entropy | 0.68898 |
| G+C content | 0.50002 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -10.75 |
| Energy contribution | -10.49 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.668712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
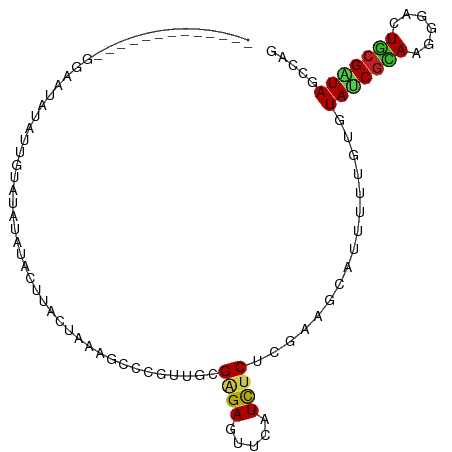
>dm3.chr2L 22105115 109 + 23011544 UAAAGCCCGUUGCGAGAGUUCAUCUCACGAAGCAUUUUUGUGUAUCGCAAGGGACUGCGAUAGCCAGGACCACCAAGCGAUGCAGAUGCACCAUUAAAAUUUGAAGAAC----------- ....((((.((((((((.....))))..((.((((....)))).)))))).)).(((((...((..((....))..))..)))))..))....................----------- ( -29.00, z-score = -1.84, R) >dp4.chr4_group1 1497763 83 + 5278887 UAAAGACCAUUGCGAGAGUUCAUCUCUUUGAGCAUCUUAUUGUAUCGCAGGGGACUGCGAUAGCCGGGUCCACCAAAGGCAGC------------------------------------- ....((((...(((((((((((......))))).))))....((((((((....))))))))))..)))).............------------------------------------- ( -31.40, z-score = -3.45, R) >droPer1.super_5 3103851 83 - 6813705 UAAAGACCAUUGCGAGAGUUCAUCUCUUUGAGCAUCUUAUUGUAUCGCAGGGGACUGCGAUAGCCGGGUCCACCAAAGGCAGC------------------------------------- ....((((...(((((((((((......))))).))))....((((((((....))))))))))..)))).............------------------------------------- ( -31.40, z-score = -3.45, R) >droAna3.scaffold_12943 2297883 109 - 5039921 UAGAGCCCGUUGCGAGAGUUCAUUUCGCGCAUGAUCUUCUGAUAACGUAGAGGACUACGAUAUCCGGGGCCACCGAGAGCAGCGGCGCCACUUUGCAUGUUCCUGGACU----------- ..(.((((((.((((((.....))))))))........(.((((.(((((....))))).)))).)))))).(((.(((((...(((......))).))))).)))...----------- ( -35.60, z-score = -0.80, R) >droEre2.scaffold_4845 2348555 109 + 22589142 UAAAGCCCGUUUCGAGAGUUCAUUUCACGAAGCAUUUUUGUGUAUCGUAAGGGACUGCGGUACCCAGGACCUCCAAGAGAUGCGGAACCGCCAUUAAAAUUUGUAGACC----------- .....(((((((((((.((((.....((((.((((....)))).))))..(((((....)).))).)))))))...)))))).))........................----------- ( -26.80, z-score = -0.91, R) >droYak2.chr2R 650498 109 + 21139217 UAAAGCCCGUUUCGAGAGUUCAUUUCACGAAGCAUUUUUGUGUAUCGUAAGGGACUGCGAUAACCAGGACCUCCGAGCGAAGCGGAUCCGCCAUUAAAAUUUGAAGACC----------- ......(((((((....((((.....((((.((((....)))).)))).(((..(((.(....))))..)))..)))))))))))........................----------- ( -26.90, z-score = -0.88, R) >droSec1.super_28 709413 109 + 873875 UAAAGCCCGUUUCGAGAGUUCAUUUCACGAAGCAUUUUUGUGUAUCGCAAGGGACUGCGAUACCCAGGACCACCAAGCGAUGCAGAUCCACCAUUAAAAUUUGAAGAUC----------- ........((((((.(((.....))).)))))).......((((((((..((..(((.(....))))..)).....))))))))((((...((........))..))))----------- ( -27.50, z-score = -2.31, R) >droSim1.chr2L 21675752 109 + 22036055 UAAAGCCCGUUUCGAGAGUUCAUUUCACGAAGCAUUUUUGUGUAUCGCAAGGGACUGCGAUACCCAGGACCACCAAGCGAUGCAGAUCCACCAUUAAAAUUUGAAGAUC----------- ........((((((.(((.....))).)))))).......((((((((..((..(((.(....))))..)).....))))))))((((...((........))..))))----------- ( -27.50, z-score = -2.31, R) >droWil1.scaffold_181071 548282 107 + 1211509 UAUAGACCGUUCCGCGAGUUCAUCUCUUUCAAUGCUUUGCUGUAUCUCAAAGGACUGCGAUAGCCGGGUCCACCAAAUGCUGCCGAACCAUUCUGACAAUUCCUAGA------------- ..((((..((((.(((((.....))).......((((((.((.....))..(((((((....))..)))))..)))).)).)).))))...))))............------------- ( -20.50, z-score = 0.63, R) >droVir3.scaffold_12963 15043540 120 - 20206255 UAGAGUCCGUUGCGCGAGUUCAUUUCUCUUAAGACUUUUGUGUAUCGCAGUGGGCUGCGGUAGCCGGGACCUCCAAAGGAAGCCGAUCCGCUCUGACAGUUCCUCGAGUCGUCAUAGGAC ....((((.....((((...(((...((....)).....)))..))))((((((...((((..((.((....))...))..)))).)))))).((((..(((...)))..))))..)))) ( -34.70, z-score = 0.74, R) >droMoj3.scaffold_6500 17431122 120 + 32352404 UAGAGUCCAUUGCGGGAAUUCAUCUCUCUCAAGACCUUUGAAUACCGAAGUGGGCUGCGGUAACCAGGACCGCCAAAGGAAGCGGAGCCGCCUUGGCAGUUCCUGCCGUCGUCAUAUGAC ...((((((((.(((..(((((....((....))....))))).))).))))))))((((....((((((.(((((.((..((...))..))))))).).))))))))).(((....))) ( -40.60, z-score = -1.01, R) >droGri2.scaffold_15252 9368126 120 - 17193109 UAGAGUCCGUUGCGAGAGUUCAUCUCUCGCAUGACCUUGGUGUAACGCAGGGGGCUGCGAUAGCCGGGACCCCCGAAGGAAGCGGAUCCACUUUGGCAAUUCCUCGAGUCAUCAUAGGAC ....((((..((((((((.....))))))))((((((((((....(((((....)))))...)))))).....(((.((((((.((......)).))..))))))).)))).....)))) ( -45.90, z-score = -1.72, R) >consensus UAAAGCCCGUUGCGAGAGUUCAUCUCUCGAAGCAUUUUUGUGUAUCGCAAGGGACUGCGAUAGCCAGGACCACCAAAGGAAGCGGAUCCACCAUGAAAAUUCCUAGA_C___________ ......((.....((((.....))))................(((((((......)))))))....)).................................................... (-10.75 = -10.49 + -0.26)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:57:41 2011