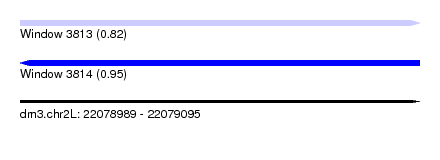
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 22,078,989 – 22,079,095 |
| Length | 106 |
| Max. P | 0.949036 |

| Location | 22,078,989 – 22,079,095 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 53.27 |
| Shannon entropy | 0.66461 |
| G+C content | 0.39064 |
| Mean single sequence MFE | -28.75 |
| Consensus MFE | -0.38 |
| Energy contribution | -0.28 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.38 |
| Mean z-score | -4.02 |
| Structure conservation index | 0.01 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.816786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
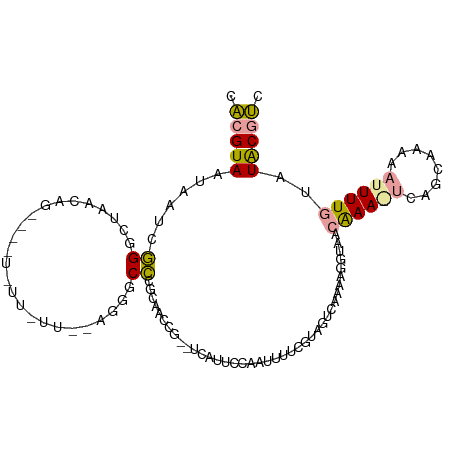
>dm3.chr2L 22078989 106 + 23011544 UACGUAUUAAUCGGGCUAACAGCUGCUUUUAUUAGAGGGCCGGCAACCG--UCCUUCCAAUUUUCGUAGUCAAAAGGUAACAAAGUCAGCAAAAAUUUUGUAUGCGUC .((((((.......(((....(((((....(((.(((((.(((...)))--.))))).)))....))))).....))).(((((((........))))))))))))). ( -23.00, z-score = -0.81, R) >droSim1.chr2L 21652324 93 + 22036055 CACGUAAUAAUCGGGCUAGCAG-------------AGCGCCUGCAGCCG--UCAUUCCAAUUUUCGUAGUCAAAAGGUAUCAAAAUCAGCAAAAAUUUUGUGUACGUC .(((((.......((((.((((-------------.....)))))))).--.............................((((((........))))))..))))). ( -17.40, z-score = -0.82, R) >droAna3.scaffold_12943 3463209 101 + 5039921 GGUGUACAAAAUUGCAUAUAAAA-AAUAUUCUUGUUCGGCACGUGACUGAUUUGUUUUUGUUUUUCUUGC----ACGUGCCGAA-CAAGAAUAUUUUUUAUAUACCC- ((((((......)))((((((((-(((((((((((((((((((((...((..............))...)----))))))))))-))))))))))))))))))))).- ( -45.84, z-score = -10.41, R) >consensus CACGUAAUAAUCGGGCUAACAG____U_UU_UU__AGGGCCCGCAACCG__UCAUUCCAAUUUUCGUAGUCAAAAGGUAACAAA_UCAGCAAAAAUUUUGUAUACGUC .(((((......((.........................)).......................................((((((........))))))..))))). ( -0.38 = -0.28 + -0.10)

| Location | 22,078,989 – 22,079,095 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 53.27 |
| Shannon entropy | 0.66461 |
| G+C content | 0.39064 |
| Mean single sequence MFE | -29.05 |
| Consensus MFE | -0.52 |
| Energy contribution | -2.86 |
| Covariance contribution | 2.35 |
| Combinations/Pair | 1.40 |
| Mean z-score | -4.45 |
| Structure conservation index | 0.02 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
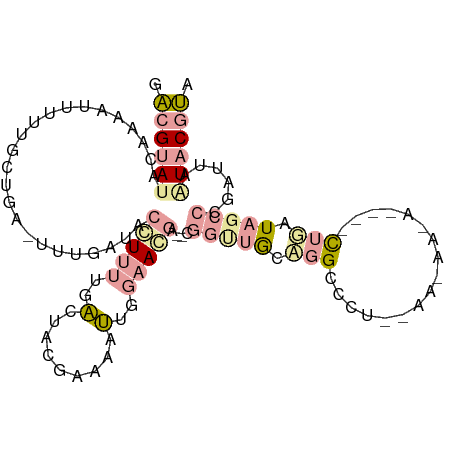
>dm3.chr2L 22078989 106 - 23011544 GACGCAUACAAAAUUUUUGCUGACUUUGUUACCUUUUGACUACGAAAAUUGGAAGGA--CGGUUGCCGGCCCUCUAAUAAAAGCAGCUGUUAGCCCGAUUAAUACGUA ..................((((((.(((((...(((((....)))))((((((.((.--(((...))).)).))))))...)))))..)))))).............. ( -20.60, z-score = -0.08, R) >droSim1.chr2L 21652324 93 - 22036055 GACGUACACAAAAUUUUUGCUGAUUUUGAUACCUUUUGACUACGAAAAUUGGAAUGA--CGGCUGCAGGCGCU-------------CUGCUAGCCCGAUUAUUACGUG .(((((..(((((((......)))))))...(((((((....)))))...)).....--.((((((((.....-------------)))).)))).......))))). ( -20.70, z-score = -1.90, R) >droAna3.scaffold_12943 3463209 101 - 5039921 -GGGUAUAUAAAAAAUAUUCUUG-UUCGGCACGU----GCAAGAAAAACAAAAACAAAUCAGUCACGUGCCGAACAAGAAUAUU-UUUUAUAUGCAAUUUUGUACACC -..((((((((((((((((((((-((((((((((----(...((..............))...)))))))))))))))))))))-))))))))))............. ( -45.84, z-score = -11.37, R) >consensus GACGUAUACAAAAUUUUUGCUGA_UUUGAUACCUUUUGACUACGAAAAUUGGAACGA__CGGUUGCAGGCCCU__AA_AA_A____CUGAUAGCCCGAUUAAUACGUA .((((((........................(((((..(.........)..)))))....(((((.(((.................))).)))))......)))))). ( -0.52 = -2.86 + 2.35)

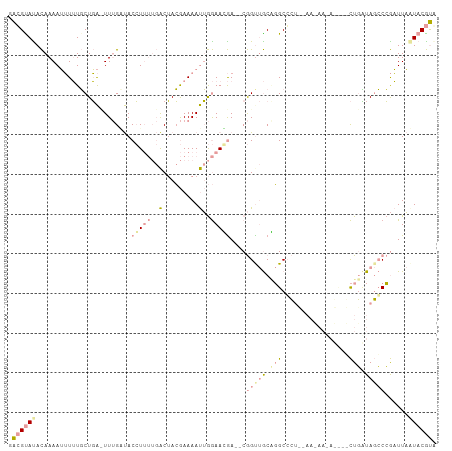
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:57:39 2011