
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 22,035,555 – 22,035,650 |
| Length | 95 |
| Max. P | 0.825750 |

| Location | 22,035,555 – 22,035,650 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.55 |
| Shannon entropy | 0.42956 |
| G+C content | 0.50692 |
| Mean single sequence MFE | -35.62 |
| Consensus MFE | -22.86 |
| Energy contribution | -22.39 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.825750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 22035555 95 - 23011544 --------------------GGUUCUC-CAAAAGGGUGGGUGGU-CGUGCGUAGGUCGACAUGUUUGGCGUAUUGUCGAAUUGCAUAAUAUGCACUUUUUCAUCGGCCAUCCGCCAA --------------------((....)-).....((((((((((-((((...(((((((((............))))))..(((((...))))))))...)).)))))))))))).. ( -34.60, z-score = -2.92, R) >droSim1.chr2L 21598484 112 - 22036055 --GGUUCUCUGGUGAAGGGGUGGAUGC-CAAAAGCGG-GGUGGU-CGUGCGUAGGUCGACAUGUUUGGCGUAUUGUCGAAUUGCAUAAUAUGCACUUUUUCAUCGGCCAUCCGCCAA --((....((((((((((((((.(((.-.....(((.-(.....-).)))((((.((((((............)))))).))))....))).))))))))))))))....))..... ( -35.60, z-score = -0.40, R) >droSec1.super_28 634751 112 - 873875 --GGUUCUCUGGUGAAGGGGUGGAUGC-CAAAAGCGG-GGUGGU-CGUGCGUAGGUCGACAUGUUUGGCGUAUUGUCGAAUUGCAUAAUAUGCACUUUUUCAUCGGCCAUCCGCCAA --((....((((((((((((((.(((.-.....(((.-(.....-).)))((((.((((((............)))))).))))....))).))))))))))))))....))..... ( -35.60, z-score = -0.40, R) >droYak2.chr2R 20358652 106 + 21139217 -------UCUGGUAAAGGGAUGGAUUU-CAAAA-GGG-GGUGGU-CAUGCGUAGGUCGACAUGUUUGGCGUAUUGCCGAAUUGCAUAAUAUGCACUUUUUCAUCGGCCAUCCGCCAA -------..((((...(..(....)..-)....-..(-((((((-((((...((((......(((((((.....))))))).((((...))))))))...))).)))))))))))). ( -30.40, z-score = -0.44, R) >droEre2.scaffold_4845 20116214 104 + 22589142 -------UCUGGUAACGGGAUGGCUUC-CAAA---GG-GGUGGU-UGUGCGUAGGUCGACAUGUUUGGCGUAUUGUCGAAUUGCAUAAUAUGCACUUUUUCAUCGGCCAUCCGCCAA -------..((((....((((((((..-..((---((-((((((-((((((....((((((............))))))..))))))))...))))))))....)))))))))))). ( -38.30, z-score = -2.62, R) >droAna3.scaffold_12943 2247708 113 + 5039921 GGAUGACUGGUGGGUGGUGGCGAAUUU-CAGGC-AGU-GGUGGC-UGUGCGUAGGUCGACAUGUUUGGCGUAUUGUCGAAUUGCAUAAUAUGCACUUUUUCAUCGGCCAUCCGCCAA .......(((((((((((.(.(((...-(((((-(.(-(.((((-(.......))))).))))))))(((((((((........))))))))).....)))..).))))))))))). ( -39.60, z-score = -1.23, R) >dp4.chr4_group1 1434146 101 - 5278887 ----------GGGGGGGGGGGGGCCAU-UGGG---GU-GGCGGU-UAUGCGUAGGUCGACAUGUUUGGCGUAUUGUCGAAUUGCAUAAUAUGCACUUUUUCAUCGGCCAUCCGCCAA ----------....((.((..((((..-((..---(.-.(((((-((((((....((((((............))))))..)))))))).)))....)..))..))))..)).)).. ( -39.40, z-score = -2.54, R) >droPer1.super_5 3041890 95 + 6813705 ----------------GGGGGGGGCAU-UGGG---GU-GGCGGU-UAUGCGUAGGUCGACAUGUUUGGCGUAUUGUCGAAUUGCAUAAUAUGCACUUUUUCAUCGGCCAUCCGCCAA ----------------((.((((((..-((..---(.-.(((((-((((((....((((((............))))))..)))))))).)))....)..))...))).))).)).. ( -35.40, z-score = -2.36, R) >droVir3.scaffold_12963 14979407 103 + 20206255 -------------GCGAGUGUGAGUGUGUGUGGUUGUGUGUGGUGCGUACGUAGGUCGAUAUGUUUGGCGUAUUGUCGAAUUGCAUAAUAUGCACUUUUUCAUCGGCCAUCCGCAA- -------------...........((((..((((((.(((.((((((((.((((.((((((............)))))).))))....))))))))....)))))))))..)))).- ( -31.70, z-score = -0.53, R) >consensus _______U__GGUGAAGGGGUGGAUUU_CAAAA__GU_GGUGGU_CGUGCGUAGGUCGACAUGUUUGGCGUAUUGUCGAAUUGCAUAAUAUGCACUUUUUCAUCGGCCAUCCGCCAA ......................................(((((..........((((((...(((((((.....)))))))(((((...)))))........))))))..))))).. (-22.86 = -22.39 + -0.47)

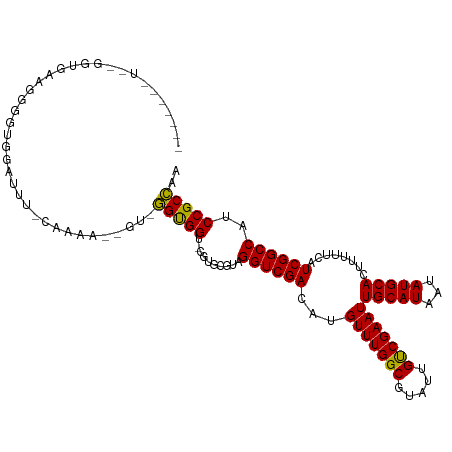
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:57:32 2011