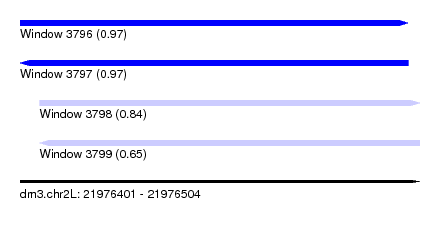
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,976,401 – 21,976,504 |
| Length | 103 |
| Max. P | 0.972541 |

| Location | 21,976,401 – 21,976,501 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 84.12 |
| Shannon entropy | 0.32328 |
| G+C content | 0.42460 |
| Mean single sequence MFE | -22.94 |
| Consensus MFE | -11.85 |
| Energy contribution | -12.08 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.971943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
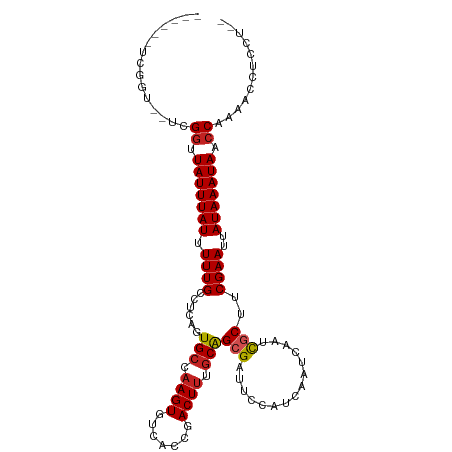
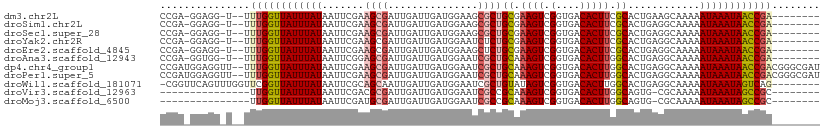
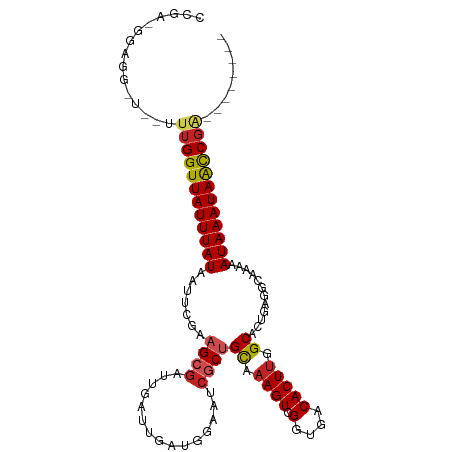
>dm3.chr2L 21976401 100 + 23011544 ------UCGGU--UCGGUUAUUUAUUUUUGCUUCAGUGCGAAGUGUCACCGACUUCGCAGCGCUUCCAUCAAUCAAUCGCUUCGAAUUAUAAAUAACCAAAACCUCCU-- ------..(((--(.((((((((((.((((......((((((((.......))))))))(((...............)))..))))..))))))))))..))))....-- ( -26.16, z-score = -3.98, R) >droSim1.chr2L 21549962 100 + 22036055 ------UCGGU--UCGGUUAUUUAUUUUUGCCUCAGUGCGAAGUGUCACCGACUUCGCAGCGCUUCCAUCAAUCAAUCGCUUCGAAUUAUAAAUAACCAAAACCUCCU-- ------..(((--(.((((((((((.((((......((((((((.......))))))))(((...............)))..))))..))))))))))..))))....-- ( -26.16, z-score = -4.04, R) >droSec1.super_28 584676 100 + 873875 ------UCGGU--UCGGUUAUUUAUUUUUGCCUCAGUGCGAAGUGUCACCGACUUCGCAGCGCUUCCAUCAAUCAAUCGCUUCGAAUUAUAAAUAACCAAAACCUCCU-- ------..(((--(.((((((((((.((((......((((((((.......))))))))(((...............)))..))))..))))))))))..))))....-- ( -26.16, z-score = -4.04, R) >droYak2.chr2R 20302106 100 - 21139217 ------UCGGU--UCGGUUAUUUAUUUUUGCCUCAGUGCGAAGUGUCACCGACUUCGCAGAGAUUCCAUCAAUCAAUCGCUUCGAAUUAUAAAUAACCAAAACCUCCU-- ------..(((--(.((((((((((.((((......((((((((.......))))))))..((((.....))))........))))..))))))))))..))))....-- ( -26.20, z-score = -3.85, R) >droEre2.scaffold_4845 20058342 100 - 22589142 ------UCGGU--UCGGUUAUUUAUUUUUGCCUCAGUGCGAAGUGUCACCGACUUCGCAGAGCUUCCAUCAAUCAAUCGCUUCGAAUUAUAAAUAACCAAAACCUCCU-- ------..(((--(.((((((((((.((((......((((((((.......))))))))((((...............))))))))..))))))))))..))))....-- ( -26.56, z-score = -4.13, R) >droAna3.scaffold_12943 2200846 100 - 5039921 ------UCGCU--UCGGUUAUUUAUUUUUGCCUCAGUGCCAAGUGUCACCGACUUUGCAGCGAUUCCAUCAAUCAAUCGCUCCGAAUUAUAAAUAACCAAAACCACCU-- ------.....--..((((((((((.((((......(((.((((.......)))).)))((((((.........))))))..))))..))))))))))..........-- ( -22.20, z-score = -3.73, R) >dp4.chr4_group1 1381496 110 + 5278887 UCGCCAUCGCCCGUCGGUUAUUUAUUUUUGCCUCAGUGCCAAGUGUCACCGACUUUGCAGCGAUUCCAUCAAUCAAUCGCUUCGAAUUAUAAAUAACCAAAAACCUCCAU ...............((((((((((.((((......(((.((((.......)))).)))((((((.........))))))..))))..))))))))))............ ( -22.00, z-score = -2.81, R) >droPer1.super_5 2988615 110 - 6813705 UCGCCAUCGCCCGUCGGUUAUUUAUUUUUGCCUCAGUGCCAAGUGUCACCGACUUUGCAGCGAUUCCAUCAAUCAAUCGCUUCGAAUUAUAAAUAACCAAAAACCUCCAU ...............((((((((((.((((......(((.((((.......)))).)))((((((.........))))))..))))..))))))))))............ ( -22.00, z-score = -2.81, R) >droWil1.scaffold_181071 455911 96 + 1211509 -------------CUGACUAUUUAUUUUUGCCUCAGUGCCAAGUGUCACCGACUAUACAGCGAUUCCAUCAAUCAAUUGCUGCGAAUUAUAAAUAACCGAACCAAACUG- -------------.....(((((((.(((((.((.(((.(....).))).))......(((((((.........))))))))))))..)))))))..............- ( -14.10, z-score = -1.05, R) >droVir3.scaffold_12963 14925884 86 - 20206255 -------------GCGGCUAUUUAUUUUUGCG-CACUGCCAAGUGUCACCGACUUUGCGGCGAUUCCAUCAAUCAAUCGCGUCGAAUUAUAAAUAACCAA---------- -------------..((.(((((((.((((((-(.((((.((((.......)))).)))).((((.....))))....))).))))..))))))).))..---------- ( -22.10, z-score = -1.93, R) >droMoj3.scaffold_6500 17308755 86 + 32352404 -------------GCGGCUAUUUAUUUUUGCG-CACUGCCAAGUGUCACCGACUUUGCGGCGAUUCCAUCAAUCAAUCGCAUCGAAUUAUAAAUAACCAA---------- -------------..((.(((((((..((..(-((((....)))))...(((...(((((.((((.....))))..))))))))))..))))))).))..---------- ( -18.70, z-score = -1.12, R) >consensus ______UCGGU__UCGGUUAUUUAUUUUUGCCUCAGUGCCAAGUGUCACCGACUUUGCAGCGAUUCCAUCAAUCAAUCGCUUCGAAUUAUAAAUAACCAAAACCUCCU__ ...............((.(((((((.((((......(((.((((.......)))).)))(((...............)))..))))..))))))).))............ (-11.85 = -12.08 + 0.22)

| Location | 21,976,401 – 21,976,501 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 84.12 |
| Shannon entropy | 0.32328 |
| G+C content | 0.42460 |
| Mean single sequence MFE | -29.53 |
| Consensus MFE | -18.51 |
| Energy contribution | -19.10 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.972541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 21976401 100 - 23011544 --AGGAGGUUUUGGUUAUUUAUAAUUCGAAGCGAUUGAUUGAUGGAAGCGCUGCGAAGUCGGUGACACUUCGCACUGAAGCAAAAAUAAAUAACCGA--ACCGA------ --....(((((.((((((((((..((((.((((...............))))(((((((.(....))))))))..))))......))))))))))))--)))..------ ( -32.06, z-score = -3.58, R) >droSim1.chr2L 21549962 100 - 22036055 --AGGAGGUUUUGGUUAUUUAUAAUUCGAAGCGAUUGAUUGAUGGAAGCGCUGCGAAGUCGGUGACACUUCGCACUGAGGCAAAAAUAAAUAACCGA--ACCGA------ --....(((((.((((((((((.......((((...............))))(((((((.(....))))))))............))))))))))))--)))..------ ( -32.06, z-score = -3.40, R) >droSec1.super_28 584676 100 - 873875 --AGGAGGUUUUGGUUAUUUAUAAUUCGAAGCGAUUGAUUGAUGGAAGCGCUGCGAAGUCGGUGACACUUCGCACUGAGGCAAAAAUAAAUAACCGA--ACCGA------ --....(((((.((((((((((.......((((...............))))(((((((.(....))))))))............))))))))))))--)))..------ ( -32.06, z-score = -3.40, R) >droYak2.chr2R 20302106 100 + 21139217 --AGGAGGUUUUGGUUAUUUAUAAUUCGAAGCGAUUGAUUGAUGGAAUCUCUGCGAAGUCGGUGACACUUCGCACUGAGGCAAAAAUAAAUAACCGA--ACCGA------ --....(((((.((((((((((...((((.....))))......(..((..((((((((.(....)))))))))..))..)....))))))))))))--)))..------ ( -33.50, z-score = -4.03, R) >droEre2.scaffold_4845 20058342 100 + 22589142 --AGGAGGUUUUGGUUAUUUAUAAUUCGAAGCGAUUGAUUGAUGGAAGCUCUGCGAAGUCGGUGACACUUCGCACUGAGGCAAAAAUAAAUAACCGA--ACCGA------ --....(((((.((((((((((...((((.....))))..........(((((((((((.(....)))))))))..)))......))))))))))))--)))..------ ( -32.50, z-score = -3.64, R) >droAna3.scaffold_12943 2200846 100 + 5039921 --AGGUGGUUUUGGUUAUUUAUAAUUCGGAGCGAUUGAUUGAUGGAAUCGCUGCAAAGUCGGUGACACUUGGCACUGAGGCAAAAAUAAAUAACCGA--AGCGA------ --.....(((((((((((((((..((((((((((((.........)))))))((.((((.(....))))).)).)))))......))))))))))))--)))..------ ( -34.80, z-score = -4.55, R) >dp4.chr4_group1 1381496 110 - 5278887 AUGGAGGUUUUUGGUUAUUUAUAAUUCGAAGCGAUUGAUUGAUGGAAUCGCUGCAAAGUCGGUGACACUUGGCACUGAGGCAAAAAUAAAUAACCGACGGGCGAUGGCGA .....(.(..((((((((((((.......(((((((.........)))))))((....((((((.(....).)))))).))....))))))))))))..).)........ ( -29.10, z-score = -1.55, R) >droPer1.super_5 2988615 110 + 6813705 AUGGAGGUUUUUGGUUAUUUAUAAUUCGAAGCGAUUGAUUGAUGGAAUCGCUGCAAAGUCGGUGACACUUGGCACUGAGGCAAAAAUAAAUAACCGACGGGCGAUGGCGA .....(.(..((((((((((((.......(((((((.........)))))))((....((((((.(....).)))))).))....))))))))))))..).)........ ( -29.10, z-score = -1.55, R) >droWil1.scaffold_181071 455911 96 - 1211509 -CAGUUUGGUUCGGUUAUUUAUAAUUCGCAGCAAUUGAUUGAUGGAAUCGCUGUAUAGUCGGUGACACUUGGCACUGAGGCAAAAAUAAAUAGUCAG------------- -...........(..(((((((.....(((((....((((.....)))))))))....((((((.(....).)))))).......)))))))..)..------------- ( -19.60, z-score = -0.11, R) >droVir3.scaffold_12963 14925884 86 + 20206255 ----------UUGGUUAUUUAUAAUUCGACGCGAUUGAUUGAUGGAAUCGCCGCAAAGUCGGUGACACUUGGCAGUG-CGCAAAAAUAAAUAGCCGC------------- ----------..((((((((((....(((((((..(((((.....))))).)))...))))(((.((((....))))-)))....))))))))))..------------- ( -25.60, z-score = -1.74, R) >droMoj3.scaffold_6500 17308755 86 - 32352404 ----------UUGGUUAUUUAUAAUUCGAUGCGAUUGAUUGAUGGAAUCGCCGCAAAGUCGGUGACACUUGGCAGUG-CGCAAAAAUAAAUAGCCGC------------- ----------..((((((((((.......((((((((.(..(.(...((((((......))))))..))..))))).-))))...))))))))))..------------- ( -24.50, z-score = -1.49, R) >consensus __AGGAGGUUUUGGUUAUUUAUAAUUCGAAGCGAUUGAUUGAUGGAAUCGCUGCAAAGUCGGUGACACUUGGCACUGAGGCAAAAAUAAAUAACCGA__ACCGA______ ......((..((((((((((((.......((((...............))))((.((((.(....))))).))............))))))))))))...))........ (-18.51 = -19.10 + 0.59)

| Location | 21,976,406 – 21,976,504 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 85.90 |
| Shannon entropy | 0.28056 |
| G+C content | 0.43000 |
| Mean single sequence MFE | -21.67 |
| Consensus MFE | -11.85 |
| Energy contribution | -12.08 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.844430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 21976406 98 + 23011544 --------UCGGUUAUUUAUUUUUGCUUCAGUGCGAAGUGUCACCGACUUCGCAGCGCUUCCAUCAAUCAAUCGCUUCGAAUUAUAAAUAACCAAA--A-CCUCC-UCGG --------..((((((((((.((((......((((((((.......))))))))(((...............)))..))))..))))))))))...--.-.....-.... ( -23.36, z-score = -3.26, R) >droSim1.chr2L 21549967 98 + 22036055 --------UCGGUUAUUUAUUUUUGCCUCAGUGCGAAGUGUCACCGACUUCGCAGCGCUUCCAUCAAUCAAUCGCUUCGAAUUAUAAAUAACCAAA--A-CCUCC-UCGG --------..((((((((((.((((......((((((((.......))))))))(((...............)))..))))..))))))))))...--.-.....-.... ( -23.36, z-score = -3.32, R) >droSec1.super_28 584681 98 + 873875 --------UCGGUUAUUUAUUUUUGCCUCAGUGCGAAGUGUCACCGACUUCGCAGCGCUUCCAUCAAUCAAUCGCUUCGAAUUAUAAAUAACCAAA--A-CCUCC-UCGG --------..((((((((((.((((......((((((((.......))))))))(((...............)))..))))..))))))))))...--.-.....-.... ( -23.36, z-score = -3.32, R) >droYak2.chr2R 20302111 98 - 21139217 --------UCGGUUAUUUAUUUUUGCCUCAGUGCGAAGUGUCACCGACUUCGCAGAGAUUCCAUCAAUCAAUCGCUUCGAAUUAUAAAUAACCAAA--A-CCUCC-UCGG --------..((((((((((.((((......((((((((.......))))))))..((((.....))))........))))..))))))))))...--.-.....-.... ( -23.40, z-score = -3.15, R) >droEre2.scaffold_4845 20058347 98 - 22589142 --------UCGGUUAUUUAUUUUUGCCUCAGUGCGAAGUGUCACCGACUUCGCAGAGCUUCCAUCAAUCAAUCGCUUCGAAUUAUAAAUAACCAAA--A-CCUCC-UCGG --------..((((((((((.((((......((((((((.......))))))))((((...............))))))))..))))))))))...--.-.....-.... ( -23.76, z-score = -3.41, R) >droAna3.scaffold_12943 2200851 98 - 5039921 --------UCGGUUAUUUAUUUUUGCCUCAGUGCCAAGUGUCACCGACUUUGCAGCGAUUCCAUCAAUCAAUCGCUCCGAAUUAUAAAUAACCAAA--A-CCACC-UCGG --------..((((((((((.((((......(((.((((.......)))).)))((((((.........))))))..))))..))))))))))...--.-.....-.... ( -22.20, z-score = -3.42, R) >dp4.chr4_group1 1381501 108 + 5278887 AUCGCCCGUCGGUUAUUUAUUUUUGCCUCAGUGCCAAGUGUCACCGACUUUGCAGCGAUUCCAUCAAUCAAUCGCUUCGAAUUAUAAAUAACCAAA--AACCUCCAUCGG ..........((((((((((.((((......(((.((((.......)))).)))((((((.........))))))..))))..))))))))))...--............ ( -22.00, z-score = -2.42, R) >droPer1.super_5 2988620 108 - 6813705 AUCGCCCGUCGGUUAUUUAUUUUUGCCUCAGUGCCAAGUGUCACCGACUUUGCAGCGAUUCCAUCAAUCAAUCGCUUCGAAUUAUAAAUAACCAAA--AACCUCCAUCGG ..........((((((((((.((((......(((.((((.......)))).)))((((((.........))))))..))))..))))))))))...--............ ( -22.00, z-score = -2.42, R) >droWil1.scaffold_181071 455911 101 + 1211509 --------CUGACUAUUUAUUUUUGCCUCAGUGCCAAGUGUCACCGACUAUACAGCGAUUCCAUCAAUCAAUUGCUGCGAAUUAUAAAUAACCGAACCAAACUGAACCG- --------.....(((((((.(((((.((.(((.(....).))).))......(((((((.........))))))))))))..)))))))...................- ( -14.10, z-score = -0.84, R) >droVir3.scaffold_12963 14925884 86 - 20206255 --------GCGGCUAUUUAUUUUUGCG-CACUGCCAAGUGUCACCGACUUUGCGGCGAUUCCAUCAAUCAAUCGCGUCGAAUUAUAAAUAACCAA--------------- --------..((.(((((((.((((((-(.((((.((((.......)))).)))).((((.....))))....))).))))..))))))).))..--------------- ( -22.10, z-score = -1.93, R) >droMoj3.scaffold_6500 17308755 86 + 32352404 --------GCGGCUAUUUAUUUUUGCG-CACUGCCAAGUGUCACCGACUUUGCGGCGAUUCCAUCAAUCAAUCGCAUCGAAUUAUAAAUAACCAA--------------- --------..((.(((((((..((..(-((((....)))))...(((...(((((.((((.....))))..))))))))))..))))))).))..--------------- ( -18.70, z-score = -1.12, R) >consensus ________UCGGUUAUUUAUUUUUGCCUCAGUGCCAAGUGUCACCGACUUUGCAGCGAUUCCAUCAAUCAAUCGCUUCGAAUUAUAAAUAACCAAA__A_CCUCC_UCGG ..........((.(((((((.((((......(((.((((.......)))).)))(((...............)))..))))..))))))).))................. (-11.85 = -12.08 + 0.22)
| Location | 21,976,406 – 21,976,504 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 85.90 |
| Shannon entropy | 0.28056 |
| G+C content | 0.43000 |
| Mean single sequence MFE | -27.21 |
| Consensus MFE | -18.36 |
| Energy contribution | -18.75 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.645735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
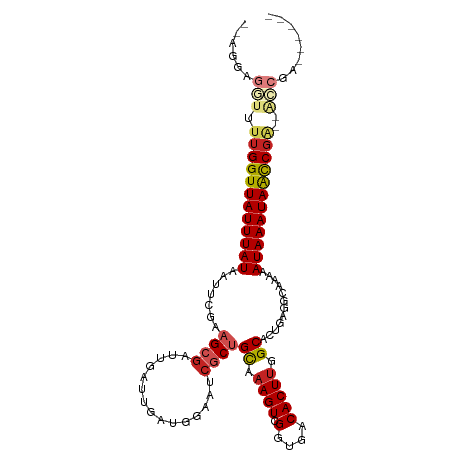
>dm3.chr2L 21976406 98 - 23011544 CCGA-GGAGG-U--UUUGGUUAUUUAUAAUUCGAAGCGAUUGAUUGAUGGAAGCGCUGCGAAGUCGGUGACACUUCGCACUGAAGCAAAAAUAAAUAACCGA-------- ((..-...))-.--.((((((((((((..((((.((((...............))))(((((((.(....))))))))..))))......))))))))))))-------- ( -27.76, z-score = -2.18, R) >droSim1.chr2L 21549967 98 - 22036055 CCGA-GGAGG-U--UUUGGUUAUUUAUAAUUCGAAGCGAUUGAUUGAUGGAAGCGCUGCGAAGUCGGUGACACUUCGCACUGAGGCAAAAAUAAAUAACCGA-------- ((..-...))-.--.((((((((((((.......((((...............))))(((((((.(....))))))))............))))))))))))-------- ( -27.76, z-score = -1.98, R) >droSec1.super_28 584681 98 - 873875 CCGA-GGAGG-U--UUUGGUUAUUUAUAAUUCGAAGCGAUUGAUUGAUGGAAGCGCUGCGAAGUCGGUGACACUUCGCACUGAGGCAAAAAUAAAUAACCGA-------- ((..-...))-.--.((((((((((((.......((((...............))))(((((((.(....))))))))............))))))))))))-------- ( -27.76, z-score = -1.98, R) >droYak2.chr2R 20302111 98 + 21139217 CCGA-GGAGG-U--UUUGGUUAUUUAUAAUUCGAAGCGAUUGAUUGAUGGAAUCUCUGCGAAGUCGGUGACACUUCGCACUGAGGCAAAAAUAAAUAACCGA-------- ((..-...))-.--.((((((((((((...((((.....))))......(..((..((((((((.(....)))))))))..))..)....))))))))))))-------- ( -29.20, z-score = -2.64, R) >droEre2.scaffold_4845 20058347 98 + 22589142 CCGA-GGAGG-U--UUUGGUUAUUUAUAAUUCGAAGCGAUUGAUUGAUGGAAGCUCUGCGAAGUCGGUGACACUUCGCACUGAGGCAAAAAUAAAUAACCGA-------- ((..-...))-.--.((((((((((((...((((.....))))..........(((((((((((.(....)))))))))..)))......))))))))))))-------- ( -28.20, z-score = -2.23, R) >droAna3.scaffold_12943 2200851 98 + 5039921 CCGA-GGUGG-U--UUUGGUUAUUUAUAAUUCGGAGCGAUUGAUUGAUGGAAUCGCUGCAAAGUCGGUGACACUUGGCACUGAGGCAAAAAUAAAUAACCGA-------- ....-.....-.--.((((((((((((..((((((((((((.........)))))))((.((((.(....))))).)).)))))......))))))))))))-------- ( -28.80, z-score = -2.46, R) >dp4.chr4_group1 1381501 108 - 5278887 CCGAUGGAGGUU--UUUGGUUAUUUAUAAUUCGAAGCGAUUGAUUGAUGGAAUCGCUGCAAAGUCGGUGACACUUGGCACUGAGGCAAAAAUAAAUAACCGACGGGCGAU ........(.(.--.((((((((((((.......(((((((.........)))))))((....((((((.(....).)))))).))....))))))))))))..).)... ( -29.10, z-score = -1.76, R) >droPer1.super_5 2988620 108 + 6813705 CCGAUGGAGGUU--UUUGGUUAUUUAUAAUUCGAAGCGAUUGAUUGAUGGAAUCGCUGCAAAGUCGGUGACACUUGGCACUGAGGCAAAAAUAAAUAACCGACGGGCGAU ........(.(.--.((((((((((((.......(((((((.........)))))))((....((((((.(....).)))))).))....))))))))))))..).)... ( -29.10, z-score = -1.76, R) >droWil1.scaffold_181071 455911 101 - 1211509 -CGGUUCAGUUUGGUUCGGUUAUUUAUAAUUCGCAGCAAUUGAUUGAUGGAAUCGCUGUAUAGUCGGUGACACUUGGCACUGAGGCAAAAAUAAAUAGUCAG-------- -.(.((((((..(((((.((((.(((..............))).)))).)))))(((....(((.(....)))).))))))))).)................-------- ( -21.54, z-score = -0.30, R) >droVir3.scaffold_12963 14925884 86 + 20206255 ---------------UUGGUUAUUUAUAAUUCGACGCGAUUGAUUGAUGGAAUCGCCGCAAAGUCGGUGACACUUGGCAGUG-CGCAAAAAUAAAUAGCCGC-------- ---------------..((((((((((....(((((((..(((((.....))))).)))...))))(((.((((....))))-)))....))))))))))..-------- ( -25.60, z-score = -1.74, R) >droMoj3.scaffold_6500 17308755 86 - 32352404 ---------------UUGGUUAUUUAUAAUUCGAUGCGAUUGAUUGAUGGAAUCGCCGCAAAGUCGGUGACACUUGGCAGUG-CGCAAAAAUAAAUAGCCGC-------- ---------------..((((((((((.......((((((((.(..(.(...((((((......))))))..))..))))).-))))...))))))))))..-------- ( -24.50, z-score = -1.49, R) >consensus CCGA_GGAGG_U__UUUGGUUAUUUAUAAUUCGAAGCGAUUGAUUGAUGGAAUCGCUGCAAAGUCGGUGACACUUGGCACUGAGGCAAAAAUAAAUAACCGA________ ...............((((((((((((.......((((...............))))((.((((.(....))))).))............))))))))))))........ (-18.36 = -18.75 + 0.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:57:26 2011