| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,973,402 – 21,973,530 |
| Length | 128 |
| Max. P | 0.902186 |

| Location | 21,973,402 – 21,973,530 |
|---|---|
| Length | 128 |
| Sequences | 11 |
| Columns | 139 |
| Reading direction | reverse |
| Mean pairwise identity | 73.15 |
| Shannon entropy | 0.56009 |
| G+C content | 0.52583 |
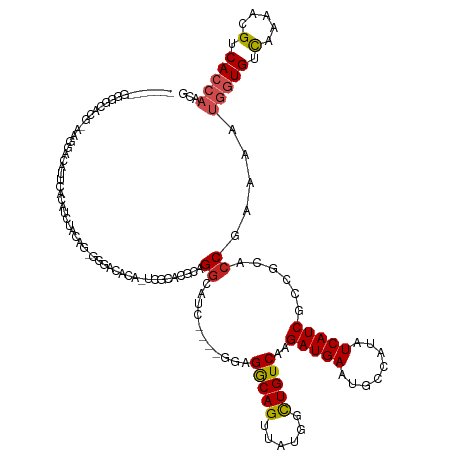
| Mean single sequence MFE | -40.66 |
| Consensus MFE | -15.96 |
| Energy contribution | -16.35 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.902186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
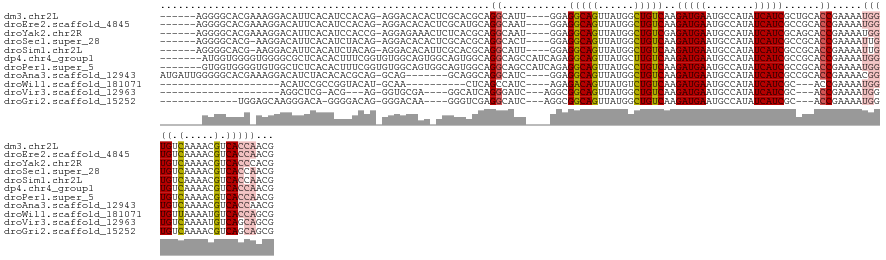
>dm3.chr2L 21973402 128 - 23011544 ------AGGGGCACGAAAGGACAUUCACAUCCACAG-AGGACACACUCGCACGCAGGCAUU----GGAGGCAGUUAUGGCUGUCAAGAUGAAUGCCAUAUCAUCGCUGCACCGAAAAUGGUGUCAAAACGUCACCAACG ------.((.((((((..((.(((((((.((((..(-((......)))((......))..)----)))((((((....))))))..).))))))))......))).))).)).....(((((.(.....).)))))... ( -39.00, z-score = -1.82, R) >droEre2.scaffold_4845 20054252 128 + 22589142 ------AGGGGCACGAAAGGACAUUCACAUCCACAG-AGGACACACUCGCAUGCAGGCAAU----GGAGGCAGUUAUGGCUGUCAAGAUGAAUGCCAUAUCAUCGCCGCACCGAAAAUGGUGUCAAAACGUCACCAACG ------.(.(((.(....)(.(((((((.((((..(-((......)))((......))..)----)))((((((....))))))..).)))))).)........))).)........(((((.(.....).)))))... ( -38.10, z-score = -1.48, R) >droYak2.chr2R 20295865 128 + 21139217 ------AGGGGCACGAAAGGACAUUCACAUCCACCG-AGGAGAAACUCUCACGCAGGCAAU----GGAGGCAGUUAUGGCUGUCGAGAUGAAUGCCAUAUCAUCGCAGCACCGAAAAUGGUGUCAAAACGUCACCCACG ------.(((..(((...((.(((((((.((((((.-.((((.....))).)...))...)----)))((((((....))))))..).))))))))...........(((((......))))).....)))..)))... ( -39.80, z-score = -1.69, R) >droSec1.super_28 581246 127 - 873875 ------AGGGGCACG-AAGGACAUUCACAUCUACAG-AGGACACACUCGCACGCAGGCACU----GGAGGCAGUUAUGGCUGUCAAGAUGAAUGCCAUAUCAUCGCCGCACCGAAAAUUGUGUCAAAACGUCACCAACG ------.(((((..(-((.....)))..........-..((((((.(((...((.(((...----...((((((....))))))..(((((........))))))))))..)))....)))))).....))).)).... ( -35.70, z-score = -1.02, R) >droSim1.chr2L 21546513 127 - 22036055 ------AGGGGCACG-AAGGACAUUCACAUCUACAG-AGGACACAUUCGCACGCAGGCAUU----GGAGGCAGUUAUGGCUGUCAAGAUGAAUGCCAUAUCAUCGCCGCACCGAAAAUUGUGUCAAAACGUCACCAACG ------.(((((..(-((.....)))..........-..((((((((((...((.(((...----...((((((....))))))..(((((........))))))))))..))))...)))))).....))).)).... ( -37.10, z-score = -1.69, R) >dp4.chr4_group1 1376347 132 - 5278887 -------AUGGUGGGGUGGGGCGCUCACACUUUCGGUGUGGCAGUGGCAGUGGCAGGCAGCCAUCAGAGGCAGUUAUGCUUGUCAAGAUGAAUGCCAUAUCAUCGCCGCACCGAAAAUGGUGUCAAAACGUCACCAACG -------.((((((.((..(((((.((...((((((((((((........((((((((((((......))).....))))))))).(((((........))))))))))))))))).)))))))...)).))))))... ( -61.60, z-score = -4.68, R) >droPer1.super_5 2983484 132 + 6813705 -------GUGGUGGGGUGUGGCUCUCACACUUUCGGUGUGGCAGUGGCAGUGGCAGGCAGCCAUCAGAGGCAGUUAUGCCUGUCAAGAUGAAUGCCAUAUCAUCGCCGCACCGAAAAUGGUGUCAAAACGUCACCAACG -------.((((((.((.((((...(.((.((((((((((((........((((((((((((......))).....))))))))).(((((........))))))))))))))))).))).))))..)).))))))... ( -61.10, z-score = -4.37, R) >droAna3.scaffold_12943 2197183 127 + 5039921 AUGAUUGGGGGCACGAAAGGACAUCUACACACGCAG-GCAG-------GCAGGCAGGCAUC----GGAGGCAGUUAUGGCUGUCAAGAUGAAUGCCAUAUCAUCGCCGCACCGAAAACGGUGUCAAAACGUCACCAACG ....((((.(((.(....)(((..........((.(-((.(-------..(((((..((((----...((((((....))))))..))))..)))).)..)...)))))((((....))))))).....))).)))).. ( -42.60, z-score = -2.20, R) >droWil1.scaffold_181071 451434 101 - 1211509 --------------------ACAUCCGCCGGUACAU-GCAA----------CUCAGCCAUC----AGAGACAGUUAUGUCUGUCAAGAUGAAUGCCAUAUCAUCGC---ACCGAAAAUGGUGUUAAAAUGUCACCAGCG --------------------.....(((.(((((((-....----------....(((((.----...(((((......)))))..(((((........)))))..---.......)))))......)))).))).))) ( -26.74, z-score = -1.93, R) >droVir3.scaffold_12963 14921448 104 + 20206255 --------------------AGGCUCG-ACG---AG-GGUGCGA----GGCAUCAGGGAUC---AGGCGGCAGUUAUGGCUGUCAAGAUGAAUGCCAUAUCAUCGC---ACCGAAAAUGGUGUCAAAAUGUCAGCAGCG --------------------..(((.(-(((---..-(((((((----(((((((......---.((((((.......))))))....)).)))))......))))---)))((........))....))))))).... ( -32.50, z-score = -0.73, R) >droGri2.scaffold_15252 9259762 114 + 17193109 -------------UGGAGCAAGGGACA-GGGGACAG-GGGACAA----GGGUCGAGGCAUC---AGGCGGCAGUUAUGGCUGUCAAGAUGAAUGCCAUAUCAUCGC---ACCGAAAAUGGUGUCAAAACGUCAGCAGCG -------------....((....(((.-(....)..-..((((.----...(((..((...---.((((((.......))))))..(((((........)))))))---..)))......)))).....)))....)). ( -33.00, z-score = -1.30, R) >consensus _______GGGGCACG_AAGGACAUUCACAUCUACAG_GGGACACA_UCGCACGCAGGCAUC____GGAGGCAGUUAUGGCUGUCAAGAUGAAUGCCAUAUCAUCGCCGCACCGAAAAUGGUGUCAAAACGUCACCAACG .......................................................((...........(((((......)))))..(((((........)))))......)).....(((((.(.....).)))))... (-15.96 = -16.35 + 0.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:57:23 2011