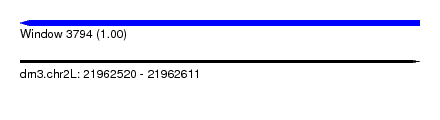
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,962,520 – 21,962,611 |
| Length | 91 |
| Max. P | 0.995113 |

| Location | 21,962,520 – 21,962,611 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 70.81 |
| Shannon entropy | 0.60698 |
| G+C content | 0.36861 |
| Mean single sequence MFE | -12.86 |
| Consensus MFE | -8.51 |
| Energy contribution | -8.41 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.995113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 21962520 91 - 23011544 GACAAA-UUC-GCAUAAUGAAAUGUUAAUUCUGACACGUCAUUAAUAUAUUUUGUUUGUCCCCAAACCA---CCAACCAUCC---AACCAUUCAACCAG ((((((-(..-...((((((..(((((....)))))..)))))).........))))))).........---..........---.............. ( -13.14, z-score = -2.84, R) >droYak2.chr2R 20283025 94 + 21139217 GACAAA-UUC-GCAUAAUGAAAUGUUAAUUUUGGCACGUCAUUAAUAUAUUUUAUUUUCCCCCAAGCCA---CCAACCACCCACAAGCCAUCCAAUCGG ......-...-...((((((..(((((....)))))..)))))).........................---........................... ( -7.90, z-score = 0.52, R) >droAna3.scaffold_12943 2186897 95 + 5039921 AACAAAUUCCAACAUAAUGAAAUGUUAAUUCUGACACGUCAUUAAUAUAUUUUGUCUGCCCUAUAGCUGCCAGUUAUCGGCUACUGGCUACCAGA---- ..............((((((..(((((....)))))..))))))......((((...(((...((((((........))))))..)))...))))---- ( -19.90, z-score = -2.06, R) >dp4.chr4_group1 1360875 89 - 5278887 GAGAAA-UUC-GUAUAAUGAAAUGUUAAUUUUGACACGUCAUUAAUAUAUUUUGUCUGGCCUCUACAAC---CAGACCAACC---CGCAACCCAACC-- ..((((-(..-((.((((((..(((((....)))))..))))))))..)))))((((((.........)---))))).....---............-- ( -16.70, z-score = -2.86, R) >droPer1.super_5 2968222 89 + 6813705 GAGAAA-UUC-GUAUAAUGAAAUGUUAAUUUUGACACGUCAUUAAUAUAUUUUGUCUGGCCUCUACAAC---CAGACCAACC---CGCAACCCAACC-- ..((((-(..-((.((((((..(((((....)))))..))))))))..)))))((((((.........)---))))).....---............-- ( -16.70, z-score = -2.86, R) >droWil1.scaffold_181071 439318 82 - 1211509 UACAAA-UUC-GCAUAAUGAAAUGUUAAUUCUGACACGUCAUUAAUAUAUUUUGUCUGCCCUCGAUG--------ACCUUUCUACAGCCAUU------- ......-...-((.((((((..(((((....)))))..)))))).........(((..........)--------)).........))....------- ( -11.30, z-score = -1.33, R) >droMoj3.scaffold_6500 17288589 86 - 32352404 CAUAAAUUUC-ACAUAAUGAAAUGUUAAUUUUGACACGUCAUUAAUAUAUUUUGUCUACCUGCCCAAUG--------CCACGCCUCCCCACCACA---- ..........-...((((((..(((((....)))))..)))))).........................--------..................---- ( -8.40, z-score = -0.86, R) >droVir3.scaffold_12963 14905787 81 + 20206255 CAUAAA-UUC-ACAUAAUGAAAUGUUAAUUUUGACACGUCAUUAAUAUAUUUUGUCUACCUUCCGAUUG--------CCACG----CCCACCACA---- .(((((-...-...((((((..(((((....)))))..))))))......)))))..............--------.....----.........---- ( -8.50, z-score = -0.90, R) >droGri2.scaffold_15252 9245811 92 + 17193109 CAUAAA-UUC-ACAUAAUGAAAUGUUAAUUUUGACACGUCAUUAAUAUAUUUUGUCUACCUUCCUGACA-ACCAUACCAACGUCCCCCCGGCACA---- ......-...-...((((((..(((((....)))))..)))))).......(((((.........))))-)..........((((....)).)).---- ( -13.20, z-score = -1.83, R) >consensus GACAAA_UUC_GCAUAAUGAAAUGUUAAUUUUGACACGUCAUUAAUAUAUUUUGUCUGCCCUCCAAACA___C__ACCAACC___CGCCACCAAA____ ..............((((((..(((((....)))))..))))))....................................................... ( -8.51 = -8.41 + -0.10)

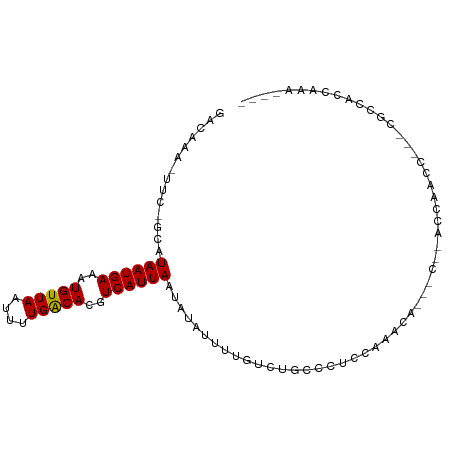
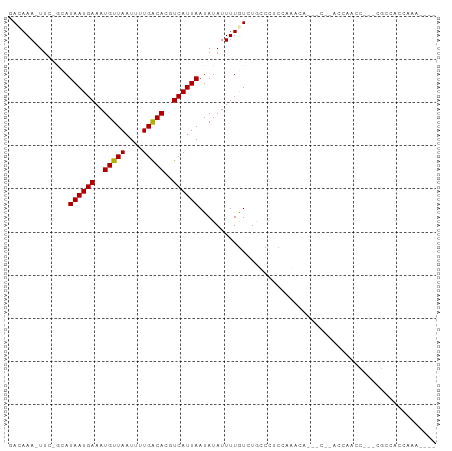
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:57:22 2011