
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,017,759 – 2,017,850 |
| Length | 91 |
| Max. P | 0.545311 |

| Location | 2,017,759 – 2,017,850 |
|---|---|
| Length | 91 |
| Sequences | 13 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 76.70 |
| Shannon entropy | 0.44134 |
| G+C content | 0.51175 |
| Mean single sequence MFE | -29.74 |
| Consensus MFE | -18.47 |
| Energy contribution | -19.00 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.545311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
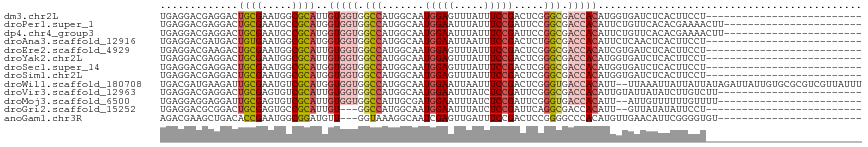
>dm3.chr2L 2017759 91 - 23011544 UGAGGACGAGGACUGCGAAUGGCGCAUUGUGGUGGCCAUGGCAAUGGAGUUUAUUUCCGACUCGGGCGACCACAUGGUGAUCUCACUUCCU-------------------------- ..((((.((((..((((.....)))).((((((.(((.(((...(((((.....)))))..)))))).))))))......))))...))))-------------------------- ( -30.70, z-score = -0.89, R) >droPer1.super_1 10250718 94 - 10282868 UGAGGACGAGGACUGCGAAUGCCGCAUGGUGGUGGCCAUGGCAAUGGAAUUUAUUUCCGAUUCCGGCGACCACAUUCUGUUCACACGAAAACUU----------------------- ...((((.((((.((((.....))))..(((((.(((..((.(((((((.....)))).)))))))).))))).))))))))............----------------------- ( -30.50, z-score = -1.64, R) >dp4.chr4_group3 8798867 94 - 11692001 UGAGGACGAGGACUGCGAAUGCCGCAUGGUGGUGGCCAUGGCAAUGGAAUUUAUUUCCGAUUCCGGCGACCACAUUCUGUUCACACGAAAACUU----------------------- ...((((.((((.((((.....))))..(((((.(((..((.(((((((.....)))).)))))))).))))).))))))))............----------------------- ( -30.50, z-score = -1.64, R) >droAna3.scaffold_12916 9733500 91 + 16180835 UGAGGACGAUGACUGUGAAUGGCGCAUGGUGGUGGCCAUGGCAAUGGAAUUAAUUUCCGACUCUGGCGACCACAUUCUCAACUCACUUCCU-------------------------- ((((((.......((((.....))))..(((((.((((.((...(((((.....))))).)).)))).))))).))))))...........-------------------------- ( -29.10, z-score = -1.80, R) >droEre2.scaffold_4929 2064045 91 - 26641161 UGAGGACGAAGACUGCGAAUGGCGCAUUGUGGUGGCCAUGGCAAUGGAGUUUAUUUCCGACUCGGGCGACCACAUCGUGAUCUCACUUCCU-------------------------- (((((((((....((((.....)))).((((((.(((.(((...(((((.....)))))..)))))).))))))))))..)))))......-------------------------- ( -31.70, z-score = -1.46, R) >droYak2.chr2L 2002893 91 - 22324452 UGAGGACGAGGACUGCGAAUGGCGCAUUGUGGUGGCCAUGGCAAUGGAGUUUAUUUCCGACUCGGGCGACCACAUGGUGAUCUCACUUCCU-------------------------- ..((((.((((..((((.....)))).((((((.(((.(((...(((((.....)))))..)))))).))))))......))))...))))-------------------------- ( -30.70, z-score = -0.89, R) >droSec1.super_14 1964483 91 - 2068291 UGAGGACGAGGACUGCGAAUGGCGCAUGGUGGUGGCCAUGGCAAUGGAGUUUAUUUCCGACUCGGGCGACCACAUGGUGAUCUCACUUCCU-------------------------- ..((((.((((..((((.....))))..(((((.(((.(((...(((((.....)))))..)))))).))))).......))))...))))-------------------------- ( -30.00, z-score = -0.44, R) >droSim1.chr2L 1987744 91 - 22036055 UGAGGACGAGGACUGCGAAUGGCGCAUGGUGGUGGCCAUGGCAAUGGAGUUUAUUUCCGACUCGGGCGACCACAUGGUGAUCUCACUUCCU-------------------------- ..((((.((((..((((.....))))..(((((.(((.(((...(((((.....)))))..)))))).))))).......))))...))))-------------------------- ( -30.00, z-score = -0.44, R) >droWil1.scaffold_180708 11664706 115 + 12563649 UGACGAUGAAGAUUGCGAAUGUCGCAUGGUGGUGGCCAUGGCAAUGGAAUUAAUUUCCGACUCGGGUGACCACAUU--UUAAAUUAUUAUUAUAGAUUAUUGUGCGCGUCGUUAUUU ((((((((.....((((.....))))..(((((.(((.(((...(((((.....)))))..)))))).)))))...--............................))))))))... ( -32.30, z-score = -1.41, R) >droVir3.scaffold_12963 7611926 93 + 20206255 UGAGGACGAGGACUGCGAGUGUCGCAUUGUGGUGGCCAUGGCAAUGGAAUUUAUCUCCGAUUCGGGCGACCACAUUGUAUUAUAUCUUGUCUU------------------------ ..(((((((((((.......)))(((.((((((.(((.(((...((((.......))))..)))))).)))))).))).......))))))))------------------------ ( -33.90, z-score = -2.63, R) >droMoj3.scaffold_6500 29335615 91 - 32352404 UGAGGAGGAGGAUUGCGAGUGUCGCAUUGUGGUGGCCAUUGCGAUGGAAUUUAUCUCCGAUUCGGGUGACCACAUU--AUUGUUUUUUGUUUU------------------------ .(((..((((((((((((..(((((......)))))..)))))))........)))))..)))((....)).....--...............------------------------ ( -25.60, z-score = -1.03, R) >droGri2.scaffold_15252 170896 86 - 17193109 UGAGGACGCGGACUGCGAGUGCCGCAUUGU---GGCCAUGGCAAUGGAAUUUAUCUCCGAUUCAGGCGACCACAUU--GUUAUAUAUUCCU-------------------------- ..((((.((((((.....)).))))..(((---((((((....))))......((.((......)).)))))))..--.........))))-------------------------- ( -24.00, z-score = -0.25, R) >anoGam1.chr3R 17598768 90 + 53272125 AGACGAAGCUGACACCGAAUGGCGGAUGUU---GGUAAAGGCAAUCGAGUUGAUUUCCGACUCCGGGGCCCACAUGUUGAACAUUCGGGGUGU------------------------ .......((..((((((.....))).))).---.))...(((..(.((((((.....)))))).)..))).((((.((((....)))).))))------------------------ ( -27.60, z-score = -0.24, R) >consensus UGAGGACGAGGACUGCGAAUGGCGCAUGGUGGUGGCCAUGGCAAUGGAAUUUAUUUCCGACUCGGGCGACCACAUUGUGAUCUCACUUCCU__________________________ .............((((.....))))..(((((.(((.......(((((.....))))).....))).)))))............................................ (-18.47 = -19.00 + 0.53)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:56 2011