| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,953,256 – 21,953,346 |
| Length | 90 |
| Max. P | 0.964805 |

| Location | 21,953,256 – 21,953,346 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 75.88 |
| Shannon entropy | 0.47707 |
| G+C content | 0.50417 |
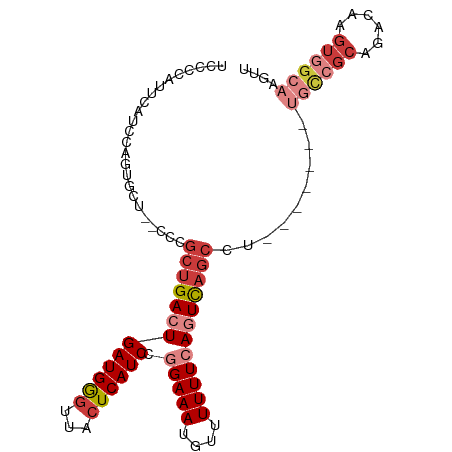
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -16.02 |
| Energy contribution | -17.81 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
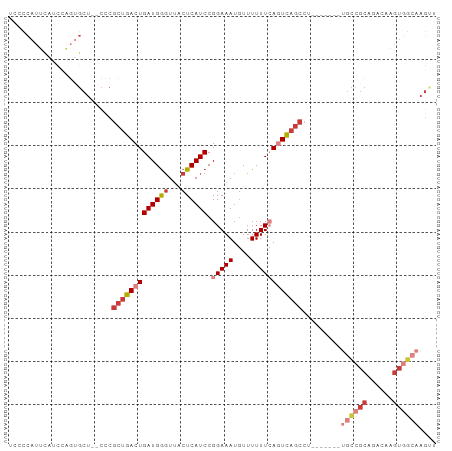
>dm3.chr2L 21953256 90 - 23011544 UUCCCAUUCAACCAGUGCU--CCCGCUGACUGAUGGGUUACUCAUCCGGAAAUGUUUUUUCAGUCAGCCUA-------GCCCCAGCCAAGUGGCAAGCU .............((((((--...(((((((((((((...)))))).(((((....))))))))))))..)-------))....(((....)))..))) ( -25.00, z-score = -1.40, R) >droAna3.scaffold_12943 2177365 99 + 5039921 UCCCCAUCUAUCCGCCGCUGGCUCGCUGACUGAUGGCUUGCUCAUCCGGAAAUGUUUUUUCACUCAGCCAGCCUCAGUACCGCAGACAAGUGGCAAGCG .............((((((..((.((.((((((.((((.(((......((((.....))))....))).))))))))).).))))...))))))..... ( -32.80, z-score = -2.04, R) >droYak2.chr2R 20273244 83 + 21139217 UCCUCAUUUACUCAGUGCU--CCCGCUGACUGAUGGGUUACUCAUCCGGAAAUUUUUUUUAAGUCAGCCU-------UGCCGCAGACAAGUC------- (((........((((((..--..))))))..((((((...)))))).)))............(((.((..-------....)).))).....------- ( -17.20, z-score = -0.33, R) >droSec1.super_28 559553 90 - 873875 UACCCAUUCAUCCAGUGCU--CCCGCUGACUGAUGGGUAACUCAUCCGGAAAUGUUUUUUCAGUCAGCCU-------UGCCGCAGACAAGUGGCAAGUU (((((((.((..(((((..--..)))))..)))))))))........(((((....))))).......((-------((((((......)))))))).. ( -30.80, z-score = -3.15, R) >droSim1.chr2L 21525326 90 - 22036055 UACCCAUUCAUCCAGUGCU--CCCGCUGACUGAUGGGUUACUCAUCCGGAAAUGUUUUUUCAGUCAGCCU-------UGCCGCAGACAAGUGGCAAGUU ...................--...(((((((((((((...)))))).(((((....))))))))))))((-------((((((......)))))))).. ( -30.40, z-score = -3.02, R) >droWil1.scaffold_181071 785497 70 + 1211509 -----------------CU--UCGCCUGACUGAUGAGUUACUCAUCCGGAAAUGUUUUUUCACUUCACUUG------UGUCG----CGAGUCGCAAGUC -----------------..--..........((((((...)))))).(((((....))))).....(((((------((.(.----...).))))))). ( -18.20, z-score = -1.39, R) >droEre2.scaffold_4845 20029561 90 + 22589142 UCACCAUUCAUCUAGUGCU--CCCGCUGACUGAUGGGUUACUCAUCCGGAAAUGUUUUUUAAGUCAGCCC-------UGCCGCAGACAAGUGGCAAGUG .(((...............--...(((((((((((((...))))))..((((....)))).)))))))..-------((((((......)))))).))) ( -24.80, z-score = -1.14, R) >consensus UCCCCAUUCAUCCAGUGCU__CCCGCUGACUGAUGGGUUACUCAUCCGGAAAUGUUUUUUCAGUCAGCCU_______UGCCGCAGACAAGUGGCAAGUU ........................(((((((((((((...)))))).(((((....))))))))))))..........(((((......)))))..... (-16.02 = -17.81 + 1.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:57:20 2011