| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,938,057 – 21,938,174 |
| Length | 117 |
| Max. P | 0.793282 |

| Location | 21,938,057 – 21,938,174 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 82.01 |
| Shannon entropy | 0.31982 |
| G+C content | 0.48358 |
| Mean single sequence MFE | -32.99 |
| Consensus MFE | -25.00 |
| Energy contribution | -26.00 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.594498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 21938057 117 + 23011544 GGACUGAGGGAGGGAAGGUCUACGGGCUUGGGCGGUGUCCUGUUAUGGCCCGCGGAAAAUUGCAGUUGAAAUUAUUGCAUUUAAACAGCACAUUAUCACA-CACAAGCCCAGAGACAC .................((((..((((((((((.(((......))).))))((.......((((((.......))))))........))...........-...))))))..)))).. ( -34.36, z-score = -1.03, R) >droYak2.chr2R 20256079 117 - 21139217 GGACUGGGAAUUGGAAGGGCCACGGGCUUGGGUGGUGUCCUUUUAUGACCCGCGGAAAAUUGCAGUUGAAAUUAUUGCAUUAAAACAGGACAUUAUCACA-CACAAGCCCAGGGACAC (..((......(((.....))).((((((((((((((((((.((.(((((...)).....((((((.......))))))))).)).)))))))))))...-..))))))).))..).. ( -34.10, z-score = -1.12, R) >droSec1.super_28 543659 116 + 873875 GGACUGAGGGAGGGAAGGUCUACGGGCUUGGGCGGUGUCCUGUUAUGGCC-GCGGAAAAUUGCAGUUGAAAUUAUUGCAUUUAAACAGGACAUUAUCACA-CACAAGCCCAGAGACAC .................((((..(((((((((..((((((((((....(.-...).....((((((.......))))))....))))))))))..))...-..)))))))..)))).. ( -38.00, z-score = -2.62, R) >droSim1.chr2L 21508847 117 + 22036055 GGACUGAGGAAGGGAAGGUCUACGGGCUUGGGCGGUGUCCUGUUAUGGCCCGCGGAAAAUUGCAGUUGAAAUUAUUGCAUUUAAACAGGACAUUAUCACA-CACAAGCCCAGAGACAC .................((((..(((((((((..((((((((((....((...)).....((((((.......))))))....))))))))))..))...-..)))))))..)))).. ( -38.30, z-score = -2.52, R) >droPer1.super_5 2936241 105 - 6813705 -------------AAACUGGCAAGGUCCUGCAGGGUGUCCUGUUGUGGCCCGCGGAAAAUUGCAGUGGAAAUUAUUGCAUUUAAACAGGACAUUAUCACAGCACAGACACAAAAACAC -------------...(((((.....((....))(((((((((((((...))).......(((((((.....)))))))....)))))))))).......)).)))............ ( -26.60, z-score = -0.21, R) >dp4.chr4_group1 1329033 105 + 5278887 -------------AAACUGGCAAGGUCCUGCAGGGUGUCCUGUUGUGGCCCGCGGAAAAUUGCAGUGGAAAUUAUUGCAUUUAAACAGGACAUUAUCACAGCACAGACACAAAAACAC -------------...(((((.....((....))(((((((((((((...))).......(((((((.....)))))))....)))))))))).......)).)))............ ( -26.60, z-score = -0.21, R) >consensus GGACUGAGG_AGGGAAGGGCCACGGGCUUGGGCGGUGUCCUGUUAUGGCCCGCGGAAAAUUGCAGUUGAAAUUAUUGCAUUUAAACAGGACAUUAUCACA_CACAAGCCCAGAGACAC .......................(((((((((..((((((((((....((...)).....((((((.......))))))....))))))))))..))......)))))))........ (-25.00 = -26.00 + 1.00)


| Location | 21,938,057 – 21,938,174 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.01 |
| Shannon entropy | 0.31982 |
| G+C content | 0.48358 |
| Mean single sequence MFE | -30.87 |
| Consensus MFE | -21.32 |
| Energy contribution | -22.02 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.793282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
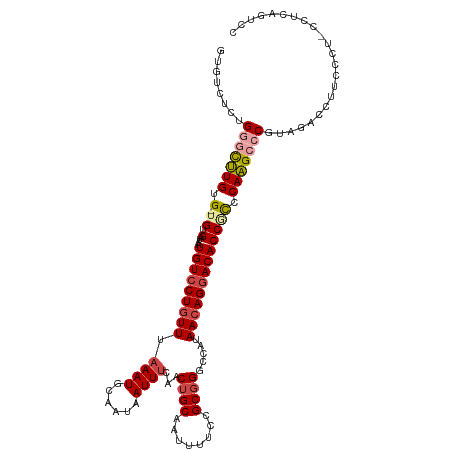
>dm3.chr2L 21938057 117 - 23011544 GUGUCUCUGGGCUUGUG-UGUGAUAAUGUGCUGUUUAAAUGCAAUAAUUUCAACUGCAAUUUUCCGCGGGCCAUAACAGGACACCGCCCAAGCCCGUAGACCUUCCCUCCCUCAGUCC ..((((.((((((((..-.(.((.(((.(((.(((.((((......)))).))).)))))).)))((((.((......))...)))).)))))))).))))................. ( -34.30, z-score = -2.41, R) >droYak2.chr2R 20256079 117 + 21139217 GUGUCCCUGGGCUUGUG-UGUGAUAAUGUCCUGUUUUAAUGCAAUAAUUUCAACUGCAAUUUUCCGCGGGUCAUAAAAGGACACCACCCAAGCCCGUGGCCCUUCCAAUUCCCAGUCC ..(..((((((((((.(-.(((....((((((((......))...........((((........))))........)))))).)))))))))))).))..)................ ( -30.80, z-score = -1.47, R) >droSec1.super_28 543659 116 - 873875 GUGUCUCUGGGCUUGUG-UGUGAUAAUGUCCUGUUUAAAUGCAAUAAUUUCAACUGCAAUUUUCCGC-GGCCAUAACAGGACACCGCCCAAGCCCGUAGACCUUCCCUCCCUCAGUCC ..((((.((((((((.(-.(((....(((((((((....(((...((((.(....).))))....))-).....))))))))).)))))))))))).))))................. ( -36.10, z-score = -4.18, R) >droSim1.chr2L 21508847 117 - 22036055 GUGUCUCUGGGCUUGUG-UGUGAUAAUGUCCUGUUUAAAUGCAAUAAUUUCAACUGCAAUUUUCCGCGGGCCAUAACAGGACACCGCCCAAGCCCGUAGACCUUCCCUUCCUCAGUCC ..((((.((((((((.(-.(((....(((((((((.((((......))))...((((........)))).....))))))))).)))))))))))).))))................. ( -37.00, z-score = -3.78, R) >droPer1.super_5 2936241 105 + 6813705 GUGUUUUUGUGUCUGUGCUGUGAUAAUGUCCUGUUUAAAUGCAAUAAUUUCCACUGCAAUUUUCCGCGGGCCACAACAGGACACCCUGCAGGACCUUGCCAGUUU------------- ......(((.((..((.((((.....(((((((((.((((......))))...((((........)))).....)))))))))....)))).))...)))))...------------- ( -23.50, z-score = 0.53, R) >dp4.chr4_group1 1329033 105 - 5278887 GUGUUUUUGUGUCUGUGCUGUGAUAAUGUCCUGUUUAAAUGCAAUAAUUUCCACUGCAAUUUUCCGCGGGCCACAACAGGACACCCUGCAGGACCUUGCCAGUUU------------- ......(((.((..((.((((.....(((((((((.((((......))))...((((........)))).....)))))))))....)))).))...)))))...------------- ( -23.50, z-score = 0.53, R) >consensus GUGUCUCUGGGCUUGUG_UGUGAUAAUGUCCUGUUUAAAUGCAAUAAUUUCAACUGCAAUUUUCCGCGGGCCAUAACAGGACACCGCCCAAGCCCGUAGACCUUCCCU_CCUCAGUCC ........(((((((....(((....(((((((((.((((......))))...((((........)))).....))))))))).))).)))))))....................... (-21.32 = -22.02 + 0.70)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:57:19 2011