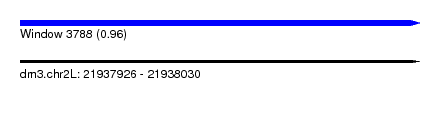
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,937,926 – 21,938,030 |
| Length | 104 |
| Max. P | 0.962039 |

| Location | 21,937,926 – 21,938,030 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.29 |
| Shannon entropy | 0.57041 |
| G+C content | 0.53523 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -15.50 |
| Energy contribution | -15.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.962039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 21937926 104 + 23011544 GGGCCA---CGUCAGUCAGCAAUAAAUAAAUGCAAGCUGGCAGUCAUUUGGUCGUCACACGUCC-UGACAGUCCGACGACCG-UGGAGGAAUGGCCACAAAGGGAAGAG----------- .(((((---.((((((..(((.........)))..))))))..((...((((((((..((((..-..)).))..))))))))-..))....))))).............----------- ( -34.20, z-score = -1.81, R) >droAna3.scaffold_12943 2163774 116 - 5039921 GGGCCA---CGUCAGUCAGCAAUAAAUAAAUGCAAGCUGGCAGUCAUUUGGUCGUCACACGGCC-AGACAGGCGGACGACUUUCGGGUGGUGGGGCAUGGCGGGGCAAUAAAGUGGAAAG ..(((.---(((((((((((...............)))))).(((.((((((((.....)))))-))).......((.(((....))).))..))).))))).))).............. ( -36.96, z-score = -1.69, R) >droEre2.scaffold_4845 20008136 104 - 22589142 GGGCCA---CGUCAGUCAGCAAUAAAUAAAUGCAAGCUGGCAGUCAUUUGGUCGUCACACGUCC-UGACAGUCGGACGACCG-UGUAGGAAUGGCCACAAAGGGAAGAG----------- .(((((---.((((((..(((.........)))..))))))..((...((((((((..((((..-..)).))..))))))))-.....)).))))).............----------- ( -33.60, z-score = -1.95, R) >droYak2.chr2R 20255954 104 - 21139217 GGGCCA---CGUCAGUCAGCAAUAAAUAAAUGCAAGCUGGCAGUCAUUUGGUCGUCACACGUCC-UGACAGUCGGACGACCG-UAGAGGAAUGGUCACAAAGGGAAGAG----------- .(((((---.((((((..(((.........)))..))))))..((...((((((((..((((..-..)).))..))))))))-.....)).))))).............----------- ( -30.90, z-score = -1.65, R) >droSec1.super_28 543528 104 + 873875 GGGCCA---CGUCAGUCAGCAAUAAAUAAAUGCAAGCUGGCAGUCAUUUGGUCGUCACACGUCC-UGACAGUCCGACGACCG-UGGAGGAAUGGCCACAAAGGGAAGAG----------- .(((((---.((((((..(((.........)))..))))))..((...((((((((..((((..-..)).))..))))))))-..))....))))).............----------- ( -34.20, z-score = -1.81, R) >droSim1.chr2L 21508716 104 + 22036055 GGGCCA---CGUCAGUCAGCAAUAAAUAAAUGCAAGCUGGCAGUCAUUUGGUCGUCACACGUCC-UGACAGUCCGACGACCG-UGGAGGAAUGGCCACAAAGGGAAGAG----------- .(((((---.((((((..(((.........)))..))))))..((...((((((((..((((..-..)).))..))))))))-..))....))))).............----------- ( -34.20, z-score = -1.81, R) >droPer1.super_5 2936116 100 - 6813705 GGGCCA---CGUCAGUCAGCAAUAAAUAAAUGCAAGCUGGCAGUCAUUUGGUCGUCACACGGGC-UAC-CAGCAGACAGCAGGCGGACGAAUGGUUGGUGGGGGG--------------- ...(((---(((((((..(((.........)))..))))))(..((((((..((((....((..-..)-).((.....)).))))..))))))..).))))....--------------- ( -34.60, z-score = -2.38, R) >dp4.chr4_group1 1328908 100 + 5278887 GGGCCA---CGUCAGUCAGCAAUAAAUAAAUGCAAGCUGGCAGUCAUUUGGUCGUCACACGGGC-CAC-CAGCAGACAGCAGGCGGACGAAUGGUUGGUGGGGGG--------------- ...(((---(((((((..(((.........)))..))))))(..((((((..((((....((..-..)-).((.....)).))))..))))))..).))))....--------------- ( -34.60, z-score = -1.99, R) >droWil1.scaffold_181071 397570 86 + 1211509 GGGCCAUCACGUCAGUCAGCAAUAAAUAAAUGCAAGCUGGCAGUCAGUUGGUCGUCACACGACC----AGACAGAACAAAAACUGAAACA------------------------------ ......(((.((((((..(((.........)))..)))))).(((...((((((.....)))))----))))...........)))....------------------------------ ( -22.60, z-score = -2.32, R) >droVir3.scaffold_12963 5340982 81 + 20206255 GGGCCA---CGUCAGUCAGCAAUAAAUAAAUGCAAGCUGGCAGUCAGUUGGUCGUCACACAGGA-CACACAC-ACACACAGGACAG---------------------------------- .(((((---(..(.((((((...............)))))).)...).)))))(((......))-)......-.............---------------------------------- ( -16.66, z-score = -0.69, R) >droMoj3.scaffold_6500 15100067 82 - 32352404 GGGCCA---CGUCAGUCAGCAAUAAAUAAAUGCAAGCUGGCAGUCAGUUGGUCGUCACACAGGA-CAUGCAGGACACACGGGGCAG---------------------------------- ..(((.---.((((((..(((.........)))..)))))).(((.....(((.(.....).))-)......)))......)))..---------------------------------- ( -21.70, z-score = -1.24, R) >droGri2.scaffold_15113 27442 91 + 133687 GGGCCA---CGUCAGUCAGCAAUAAAUAAAUGCAAGCUGGCAGUCAGUUGGUCGUCACACGGACUCGCAGGACGAACCGAAAUUGGGAAAGUGA-------------------------- ....((---(((((((..(((.........)))..))))))..(((.(((((((((...((....))...)))).)))))...)))....))).-------------------------- ( -25.50, z-score = -0.88, R) >consensus GGGCCA___CGUCAGUCAGCAAUAAAUAAAUGCAAGCUGGCAGUCAUUUGGUCGUCACACGGCC_UGACAGUCGGACGACCG_UGGAGGAAUGG_CACAAAGGGA_______________ .(((((....((((((..(((.........)))..)))))).......)))))................................................................... (-15.50 = -15.50 + 0.00)

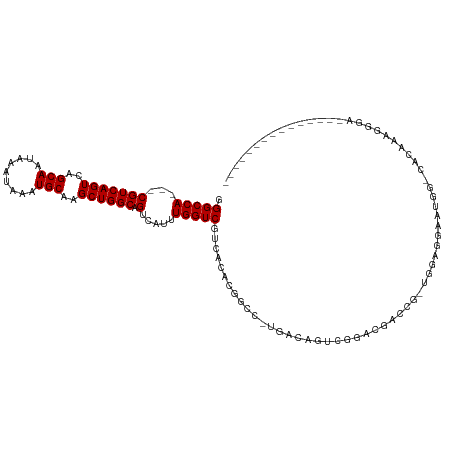
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:57:17 2011