| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,926,993 – 21,927,088 |
| Length | 95 |
| Max. P | 0.924078 |

| Location | 21,926,993 – 21,927,088 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 70.50 |
| Shannon entropy | 0.59327 |
| G+C content | 0.48333 |
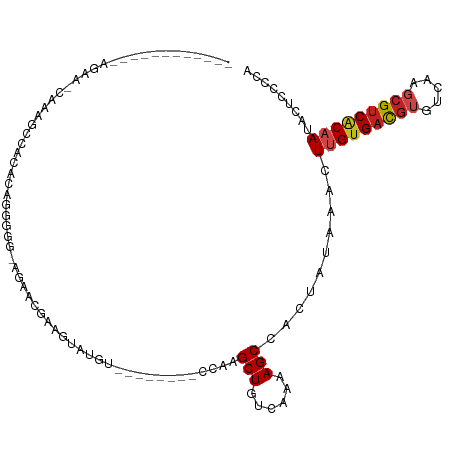
| Mean single sequence MFE | -25.16 |
| Consensus MFE | -12.20 |
| Energy contribution | -12.50 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.924078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
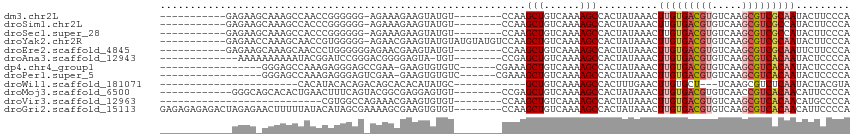
>dm3.chr2L 21926993 95 + 23011544 -----------GAGAAGCAAAGCCAACCGGGGGG-AGAAAGAAGUAUGU--------CCAAGCUGUCAAAAGCCACUAUAAACUUGUGACGUGUCAAGCGUCGCAAUACUUCCCA -----------...........((....))((((-((....((((.(((--------....(((......)))....))).))))(((((((.....)))))))....)))))). ( -26.20, z-score = -1.62, R) >droSim1.chr2L 21498708 95 + 22036055 -----------GAGAAGCAAAGCCACCCGGGGGG-AGAAAGAAGUAUGU--------CCAAGCUGUCAAAAGCCACUAUAAACUUGUGACGUGUCAAGCGUCGCCAUACUUCCCA -----------...........((....))((((-((....((((.(((--------....(((......)))....))).))))(((((((.....)))))))....)))))). ( -25.30, z-score = -1.08, R) >droSec1.super_28 533567 95 + 873875 -----------GAGAAGCAAAGCCACCCGGGGGG-AGAAAGAAGUAUGU--------CCAAGCUGUCAAAAGCCACUAUAAACUUGUGACGUGUCAAGCGUCGCCAUACUUCCCA -----------...........((....))((((-((....((((.(((--------....(((......)))....))).))))(((((((.....)))))))....)))))). ( -25.30, z-score = -1.08, R) >droYak2.chr2R 20244224 103 - 21139217 -----------GAGAACCAAAGCAACCGUGGGGG-AGAACGAAGUAUGUAUGUAUGUCCAAGCUGUCAAAAGCCACUAUAAACUUGUGACGUGUCAAGCGUCGCAAUACUUCCCA -----------...................((((-((..........((.((((.((....(((......))).)))))).))(((((((((.....)))))))))..)))))). ( -25.40, z-score = -0.78, R) >droEre2.scaffold_4845 19995215 96 - 22589142 -----------GAGAAGCAAAGCAACCCUGGGGGGAGAACGAAGUAUGU--------CCAAGCUGUCAAAAGCCACUAUAAACUUGUGACGUGUCAAGCGUCGCAAUUCUUCCCA -----------....................((((((((..((((.(((--------....(((......)))....))).))))(((((((.....)))))))..)))))))). ( -27.50, z-score = -1.49, R) >droAna3.scaffold_12943 2153617 93 - 5039921 -------------AAAAAAAAAAUACGGAUCCGGGACGGGGAGUA-UGU--------CCGAGCUGUCAAAAGCCACUAUAAACUUGUGACGUGUCAAGCGUCACAAUACUCCCCA -------------............((....))....((((((((-(..--------..(.(((......))))..........((((((((.....))))))))))))))))). ( -33.00, z-score = -4.32, R) >dp4.chr4_group1 1316261 91 + 5278887 -----------------GGGAGCCAAAGAGGGAGCCGAA-GAAGUGUGUC------CGAAAGCUGUCAAAAGCCACUAUAAACUUGUGACGUGUCAAGCGUCACAAUACUCCCCA -----------------(((((.......((..(((...-...).))..)------)....(((......)))..........(((((((((.....)))))))))..))))).. ( -26.40, z-score = -1.90, R) >droPer1.super_5 2923611 91 - 6813705 -----------------GGGAGCCAAAGAGGGAGUCGAA-GAAGUGUGUC------CGAAAGCUGUCAAAAGCCACUAUAAACUUGUGACGUGUCAAGCGUCACAAUACUCCCCA -----------------(((((.....((.((..(((..-..........------)))...)).))................(((((((((.....)))))))))..))))).. ( -26.00, z-score = -1.85, R) >droWil1.scaffold_181071 816760 77 - 1211509 -----------------------CACAUACACAGACAGCACACAUAUGC------------GCUGUCAAAAGCCACUUUGAACUUGUGCU---UCAAGCGUCUCAAUACUACGUA -----------------------..........((((((.((....)).------------))))))..((((.((.........)))))---)...((((.........)))). ( -14.70, z-score = -0.51, R) >droMoj3.scaffold_6500 15112620 95 - 32352404 ------------GGGCAGCACACUGAACUUUCAGUACGGCGAGGAGUGU--------CCGAGCUGUCAAAAGCCACUAUAAACUUGUGACGUGUCAACCGUCACAACAUUCCCCA ------------((...((..(((((....)))))...))..(((((((--------..(.(((......))))..........(((((((.(....))))))))))))))))). ( -29.90, z-score = -1.92, R) >droVir3.scaffold_12963 5353253 80 + 20206255 ---------------------------CGUGGCCAGAAACGAAGUGUGU--------CCAAGCUGUCAAAAGCCACUAUAAACUUGUGACGUGUCAAGCGUCACAACAUGCCCCA ---------------------------.(((((.........(((....--------....))).......))))).......(((((((((.....)))))))))......... ( -20.79, z-score = -1.35, R) >droGri2.scaffold_15113 38729 107 + 133687 GAGAGAGAGACUAGAGAACUUUUUAUACAUAGCGAAAAGCGAAGUGUGU--------CCAAGCUGUCAAAAGCCACUAUAAACUUGUGACGUGUCAAGCGUCACAACAUUCCCCA ...((((((.........))))))((((((.((.....))...))))))--------....(((......)))..........(((((((((.....)))))))))......... ( -21.40, z-score = -0.54, R) >consensus ____________AGAA_CAAAGCCACACAGGGGG_AGAACGAAGUAUGU________CCAAGCUGUCAAAAGCCACUAUAAACUUGUGACGUGUCAAGCGUCACAAUACUCCCCA .............................................................(((......)))..........(((((((((.....)))))))))......... (-12.20 = -12.50 + 0.30)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:57:17 2011