
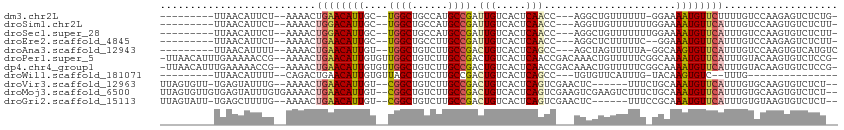
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,925,992 – 21,926,087 |
| Length | 95 |
| Max. P | 0.844678 |

| Location | 21,925,992 – 21,926,087 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 70.62 |
| Shannon entropy | 0.60388 |
| G+C content | 0.40823 |
| Mean single sequence MFE | -22.26 |
| Consensus MFE | -11.95 |
| Energy contribution | -11.78 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.27 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.781658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 21925992 95 + 23011544 ---------UUAACAUUCU--AAAACUGAACAUUGC--UGGCUGCCAUGCCGAUUGUCACUCAACC---AGGCUGUUUUUU-GGAAAAUGUUCUUUUGUCCAAGAGUCUCUG- ---------.....(((((--...((.(((((((..--((((......))))............((---(((......)))-))..)))))))....))...))))).....- ( -16.80, z-score = 0.43, R) >droSim1.chr2L 21497634 96 + 22036055 ---------UUAACAUUCU--AAAACUGGACAUUGC--UGGCUGCCAUGCCGAUUGUCACUCAACC---AGGUUGUUUUUUUGGAAAAUGUUCAUUUGUCCAAGUGUCUCUU- ---------...(((((..--..((.((((((((..--((((......))))............((---(((.......)))))..)))))))).)).....))))).....- ( -17.70, z-score = 0.47, R) >droSec1.super_28 532482 96 + 873875 ---------UUAACAUUCU--AAAACUGGACAUUGC--UGGCUGCCAUGCCGAUUGUCACUCAACC---AGGCUGUUUUUUUGGAAAAUGUUCAUUUGUCCAAGUGUCUCUU- ---------..........--......(((((((((--((((......)))(.(((.....))).)---.)))........((((((((....)))).)))))))))))...- ( -17.80, z-score = 0.49, R) >droEre2.scaffold_4845 19994140 94 - 22589142 ---------UUAACAUUCU--AAAACUGAACAUUGC--UGGCUGCCUUGCCGAUUGUCACUCAACC---AGGCUCUUUUUC--GGAAAUGUUCAUUUGUCCAAGAGUCUCUU- ---------.....(((((--..((.((((((((.(--(((.......(((..(((.....)))..---.)))......))--)).)))))))).)).....))))).....- ( -17.62, z-score = -0.32, R) >droAna3.scaffold_12943 2152679 96 - 5039921 ---------UUAACAUUUU--AAAACUGAACAUUGU--UGGCUGUCUUGCCGACUGUCACUCAGCC---AGCUAGUUUUUA-GGCAAGUGUUCAUUUGUCCAAGUGUCAUGUC ---------...((((((.--..((.((((((((((--((((......)))))).........(((---............-))).)))))))).))....))))))...... ( -23.70, z-score = -1.38, R) >droPer1.super_5 2922275 109 - 6813705 -UUAACAUUUGAAAAACCG--AAAACUGAACAUUGUGUUGGCUGUCUUGCCGACUGUCACUCAACCGACAAACUGUUUUUCGGCAAAAUGUUCAUUUGUACAAGUGUCUCCG- -...(((((((.....(((--((....(((((....((((((......))))))((((........))))...))))))))))((((.......))))..))))))).....- ( -25.10, z-score = -1.43, R) >dp4.chr4_group1 1314919 109 + 5278887 -UUAACAUUUGAAAAACCG--AAAACUGAACAUUGUGUUGGCUGUCUUGCCGACUGUCACUCAACCGACAAACUGUUUUUCGGCAAAAUGUUCAUUUGUACAAGUGUCUCCG- -...(((((((.....(((--((....(((((....((((((......))))))((((........))))...))))))))))((((.......))))..))))))).....- ( -25.10, z-score = -1.43, R) >droWil1.scaffold_181071 815832 81 - 1211509 ---------UUAACAUUUU--CAGACUGAACAUUGUGUUAGCUGUCUUGCCGACUGUCACUCAGCC---UGUGUUCAUUUG-UACAAGUGUC--UUUG--------------- ---------...((((((.--((((.(((((((.((....((.(((.....))).))......)).---.)))))))))))-...)))))).--....--------------- ( -15.90, z-score = -0.04, R) >droVir3.scaffold_12963 14848679 100 - 20206255 UUAGUGUU-UGAGUAUUUG--AAAACUGAACAUUGU--CGGCUGUCUUGCCGACUGUCACUCAGUCGAACUC------UUUCUGCAAAUGUUCAUUUGUGCAAGUGUCUCU-- ........-.(((((((((--..((.((((((((((--((((......)))))).........((.(((...------.))).)).)))))))).))...)))))).))).-- ( -29.10, z-score = -2.78, R) >droMoj3.scaffold_6500 17238071 109 + 32352404 UUAGUGUUGUGAGUAUUUGUGAAAACUGAACAUUGU--CGGCUGUCUUGCCGACUGUCACUCAGUCGAAGUCGAAGUCUUUCUGCAAAUGUUCAUUUGUGCAAGUGUCUCU-- ..........(((((((((((.(((.((((((((((--((((......))))))(((.......(((....))).........)))))))))))))).)))))))).))).-- ( -31.09, z-score = -2.17, R) >droGri2.scaffold_15113 93847 100 - 133687 UUAGUAUU-UGAGCUUUUG--AAAACUGAACAUUGU--CGGCUGUCUUGCCGACUGUCACUCAGUCGAACUC------UUUCCGCAAAUGUUCAUUUGUGUAAGUGUCUCU-- ........-.(((......--...(((((.....((--((((......))))))......)))))...((.(------((..(((((((....))))))).))).))))).-- ( -24.90, z-score = -1.86, R) >consensus _________UUAACAUUCU__AAAACUGAACAUUGU__UGGCUGUCUUGCCGACUGUCACUCAACC___AGCCUGUUUUUU_GGAAAAUGUUCAUUUGUCCAAGUGUCUCU__ ..........................((((((((....((((......)))).(((.....)))......................))))))))................... (-11.95 = -11.78 + -0.17)
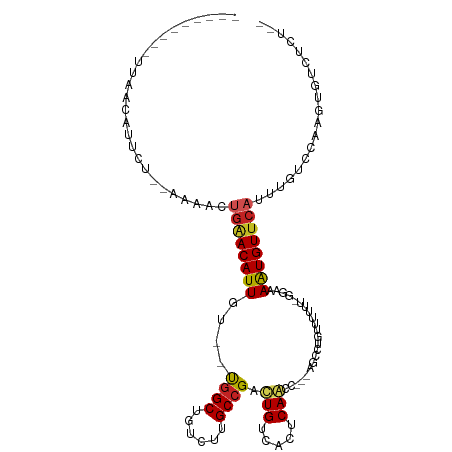

| Location | 21,925,992 – 21,926,087 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 70.62 |
| Shannon entropy | 0.60388 |
| G+C content | 0.40823 |
| Mean single sequence MFE | -22.32 |
| Consensus MFE | -14.52 |
| Energy contribution | -14.33 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.64 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.844678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 21925992 95 - 23011544 -CAGAGACUCUUGGACAAAAGAACAUUUUCC-AAAAAACAGCCU---GGUUGAGUGACAAUCGGCAUGGCAGCCA--GCAAUGUUCAGUUUU--AGAAUGUUAA--------- -..((((((.(((((.(((......))))))-))..((((..((---(((((.(((........)))..))))))--)...)))).))))))--..........--------- ( -20.10, z-score = 0.03, R) >droSim1.chr2L 21497634 96 - 22036055 -AAGAGACACUUGGACAAAUGAACAUUUUCCAAAAAAACAACCU---GGUUGAGUGACAAUCGGCAUGGCAGCCA--GCAAUGUCCAGUUUU--AGAAUGUUAA--------- -..(((((...((((((...(((....)))............((---(((((.(((........)))..))))))--)...)))))))))))--..........--------- ( -20.40, z-score = -0.39, R) >droSec1.super_28 532482 96 - 873875 -AAGAGACACUUGGACAAAUGAACAUUUUCCAAAAAAACAGCCU---GGUUGAGUGACAAUCGGCAUGGCAGCCA--GCAAUGUCCAGUUUU--AGAAUGUUAA--------- -..(((((...((((((...(((....)))..........((.(---(((((.(((........)))..))))))--))..)))))))))))--..........--------- ( -21.20, z-score = -0.36, R) >droEre2.scaffold_4845 19994140 94 + 22589142 -AAGAGACUCUUGGACAAAUGAACAUUUCC--GAAAAAGAGCCU---GGUUGAGUGACAAUCGGCAAGGCAGCCA--GCAAUGUUCAGUUUU--AGAAUGUUAA--------- -....((((((..(((...((((((((...--........((((---.(((((.......))))).)))).(...--.))))))))))))..--)))..)))..--------- ( -21.40, z-score = -0.46, R) >droAna3.scaffold_12943 2152679 96 + 5039921 GACAUGACACUUGGACAAAUGAACACUUGCC-UAAAAACUAGCU---GGCUGAGUGACAGUCGGCAAGACAGCCA--ACAAUGUUCAGUUUU--AAAAUGUUAA--------- (((((.....(((((((......((((((((-............---))).)))))...((.(((......))).--))..)))))))....--...)))))..--------- ( -23.40, z-score = -1.33, R) >droPer1.super_5 2922275 109 + 6813705 -CGGAGACACUUGUACAAAUGAACAUUUUGCCGAAAAACAGUUUGUCGGUUGAGUGACAGUCGGCAAGACAGCCAACACAAUGUUCAGUUUU--CGGUUUUUCAAAUGUUAA- -.(....).............(((((((.((((((((.....((((((..((.....))..))))))((((..........))))...))))--)))).....)))))))..- ( -24.70, z-score = -0.01, R) >dp4.chr4_group1 1314919 109 - 5278887 -CGGAGACACUUGUACAAAUGAACAUUUUGCCGAAAAACAGUUUGUCGGUUGAGUGACAGUCGGCAAGACAGCCAACACAAUGUUCAGUUUU--CGGUUUUUCAAAUGUUAA- -.(....).............(((((((.((((((((.....((((((..((.....))..))))))((((..........))))...))))--)))).....)))))))..- ( -24.70, z-score = -0.01, R) >droWil1.scaffold_181071 815832 81 + 1211509 ---------------CAAA--GACACUUGUA-CAAAUGAACACA---GGCUGAGUGACAGUCGGCAAGACAGCUAACACAAUGUUCAGUCUG--AAAAUGUUAA--------- ---------------....--((((..(((.-(....).)))((---(((((((.....((..((......))..))......)))))))))--....))))..--------- ( -17.40, z-score = -0.46, R) >droVir3.scaffold_12963 14848679 100 + 20206255 --AGAGACACUUGCACAAAUGAACAUUUGCAGAAA------GAGUUCGACUGAGUGACAGUCGGCAAGACAGCCG--ACAAUGUUCAGUUUU--CAAAUACUCA-AACACUAA --.(((..............((((.(((......)------))))))((((((..(...((((((......))))--))..)..))))))..--......))).-........ ( -23.80, z-score = -1.34, R) >droMoj3.scaffold_6500 17238071 109 - 32352404 --AGAGACACUUGCACAAAUGAACAUUUGCAGAAAGACUUCGACUUCGACUGAGUGACAGUCGGCAAGACAGCCG--ACAAUGUUCAGUUUUCACAAAUACUCACAACACUAA --.(((.....((((..(((....)))))))(((((...(((....)))((((..(...((((((......))))--))..)..))))))))).......))).......... ( -24.10, z-score = -1.13, R) >droGri2.scaffold_15113 93847 100 + 133687 --AGAGACACUUACACAAAUGAACAUUUGCGGAAA------GAGUUCGACUGAGUGACAGUCGGCAAGACAGCCG--ACAAUGUUCAGUUUU--CAAAAGCUCA-AAUACUAA --.(((.............((((((((..(((...------....(((((((.....)))))))........)))--..)))))))).....--......))).-........ ( -24.31, z-score = -1.64, R) >consensus __AGAGACACUUGGACAAAUGAACAUUUGCC_AAAAAACAGCCU___GGUUGAGUGACAGUCGGCAAGACAGCCA__ACAAUGUUCAGUUUU__AGAAUGUUAA_________ ...................((((((((....................(((((.....)))))(((......))).....)))))))).......................... (-14.52 = -14.33 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:57:16 2011