| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,876,188 – 21,876,282 |
| Length | 94 |
| Max. P | 0.917702 |

| Location | 21,876,188 – 21,876,282 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 71.17 |
| Shannon entropy | 0.58135 |
| G+C content | 0.57749 |
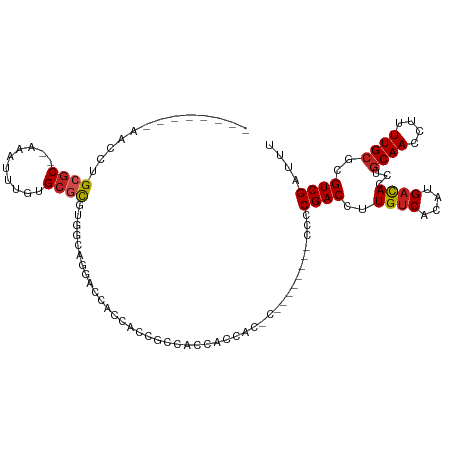
| Mean single sequence MFE | -29.32 |
| Consensus MFE | -12.43 |
| Energy contribution | -12.56 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.917702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
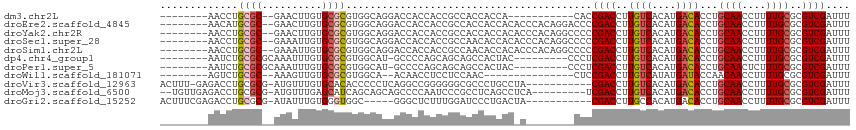
>dm3.chr2L 21876188 94 + 23011544 --------AACCUGCGC--GAACUUGUGCGCGUGGCAGGACCACCACCGCCACCACCA-----------CACCGACCUUGUCACAUGACACCUGCAACCUUUUGCGCGUCGAUUU --------.....((((--(((....((((.(((((.((.......))))))))....-----------.........((((....))))...)))....)))))))........ ( -27.20, z-score = -1.90, R) >droEre2.scaffold_4845 19941400 105 - 22589142 --------AACAUGCGC--GAACUUGUGCGCGUGGCAGGACCACCACCGCCACCACCACACCCACAGGACCCCGACCUUGUCACAUGACACCUGCAACCUUUUGCGCGUCGAUUU --------...((((((--(((..((((...(((((.((.......)))))))...))))....((((..........((((....))))))))......)))))))))...... ( -29.70, z-score = -1.84, R) >droYak2.chr2R 20186602 105 - 21139217 --------AACCUGCGC--GAACUUGUGCGCGUGGCAGGACCACCACCGCCACCACCACACCCACAGGCCCCCGACCUUGUCACAUGACACCUGCAACCUUUUGCGCGUCGAUUU --------..(((((((--(......)))))(((((.((.......)))))))............)))....((((..((((....))))...((((....))))..)))).... ( -29.30, z-score = -1.57, R) >droSec1.super_28 481932 105 + 873875 --------AACCUGCGC--GAAAUUGUGCGCGUGGCAGGACCACCACCGCCAACACCACACCCACAGGCCCCCGACCUUGUCACAUGACACCUGCAACCUUUUGCGCGUCGAUUU --------.....((((--(((((((.(((.((((........))))))))))...........((((..........((((....)))))))).....))))))))........ ( -28.10, z-score = -1.08, R) >droSim1.chr2L 21449204 105 + 22036055 --------AACCUGCGC--GAAAUUGUGCGCGUGGCAGGACCACCACCGCCAACACCACACCCACAGGCCCCCGACCUUGUCACAUGACACCUGCAACCUUUUGCGCGUCGAUUU --------.....((((--(((((((.(((.((((........))))))))))...........((((..........((((....)))))))).....))))))))........ ( -28.10, z-score = -1.08, R) >dp4.chr4_group1 1265370 97 + 5278887 --------AAUCUGCGCGCAAAUUUGUGCGCGUGGCAU-GCCCCAGCAGCAGCCACUAC---------CCCUCGACCUUGUCACAUGACACCUGCAACCUUUUGCGCGUCGAUUU --------((((.(((((((((....(((..(((((.(-((.......))))))))...---------..........((((....))))...)))....))))))))).)))). ( -36.10, z-score = -4.02, R) >droPer1.super_5 2871731 97 - 6813705 --------AAUCUGCGCGCAAAUUUGUGCGCGUGGCAU-GCCCCAGCAGCAGCCACUAC---------CCCUCGACCUUGUCACAUGACACCUGCAACUCUUUGCGCGUCGAUUU --------((((.(((((((((....(((..(((((.(-((.......))))))))...---------..........((((....))))...)))....))))))))).)))). ( -35.40, z-score = -3.67, R) >droWil1.scaffold_181071 894379 88 - 1211509 --------AGUCUGCGC--AAAGUUGUGCGCGUGGCA--ACAACCUCCUCCAAC---------------CUCCGACCUUGUCAUAUGAUACCAACAACCUUUUGCGCGUCGAUUU --------(((((((((--((((((((.((..(((((--(..............---------------........))))))..))......)))))..))))))))..)))). ( -19.35, z-score = -0.99, R) >droVir3.scaffold_12963 14786464 102 - 20206255 ACUUU-GAGACCUGCGCG-AUGUUUGUGCACACCCCCUCAGGCCGGGGGGCGCCCUGCCUA-----------CGACCUUGUCACAUGACACCUGCAACCUUUUGCGCGUCGAUUU .....-..(((..(((((-(......((((.........((((.((((....)))))))).-----------......((((....))))..)))).....)))))))))..... ( -30.50, z-score = -0.49, R) >droMoj3.scaffold_6500 17179604 103 + 32352404 --UGUUGAGACCUGCGCG-AUGUUUGAGCAUCAGCAGCAGCCCCAAUCCCGCCUCAGCCUCA---------UCGACCUUGUCACAUGACACCUGCAACCUUUUGCGCGUCGAUUU --.((((((..((((..(-((((....))))).))))..((.........))))))))...(---------(((((..((((....))))...((((....))))..)))))).. ( -29.70, z-score = -2.17, R) >droGri2.scaffold_15252 9147016 98 - 17193109 ACUUUCGAGACCUGCGCG-AUAUUUGUGGGUGGC-----GGGCUCUUUGGAUCCCUGACUA-----------CGACCUUGCCACAUGACACCUGCAACCUUUUGCGCGUCGAUUU ....((((....((((((-(...(((..((((..-----.(((.....((.((........-----------.))))..)))......))))..)))....)))))))))))... ( -29.10, z-score = -1.79, R) >consensus ________AACCUGCGC__AAAUUUGUGCGCGUGGCAGGACCACCACCGCCACCACCAC_C________CCCCGACCUUGUCACAUGACACCUGCAACCUUUUGCGCGUCGAUUU ............(((((........)))))..........................................((((..((((....))))...((((....))))..)))).... (-12.43 = -12.56 + 0.13)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:57:13 2011