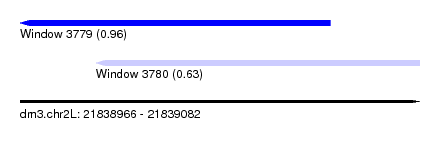
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,838,966 – 21,839,082 |
| Length | 116 |
| Max. P | 0.957139 |

| Location | 21,838,966 – 21,839,056 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 65.09 |
| Shannon entropy | 0.69535 |
| G+C content | 0.31228 |
| Mean single sequence MFE | -18.28 |
| Consensus MFE | -6.08 |
| Energy contribution | -6.15 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.957139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 21838966 90 - 23011544 ---UCUUACAUCGUAAAUCAGUCAU-U--AGCUAUCAAUUCCUA---UGAUUGAUUUACACAUUGUUUUUGAUCGUUAUUGAUCACAUUGUGUUCGUCU------------ ---.........(((((((((((((-.--..............)---))))))))))))(((.(((....(((((....)))))))).)))........------------ ( -20.96, z-score = -3.39, R) >droSim1.chr2L 21412043 90 - 22036055 ---UCUUACAUCGUAAAUCAGUCAU-C--AGCUAUCAAUUCCUA---UGAUUGAUUUACAAAUUGUUUUUGAUCGUUAUUGAUCACAUCGUGUUCGUCU------------ ---...(((...(((((((((((((-.--((..........)))---))))))))))))..........((((((....))))))....))).......------------ ( -20.30, z-score = -3.35, R) >droSec1.super_28 397583 90 - 873875 ---UCUUACAUCGUAAAUCAGUCAU-C--AGCUAUCAAAUCCUA---UGAUUGAUUUACAAAUUGUUUUUGAUCGUUAUUGAUCACAUUGUGUUCGUCU------------ ---...((((..(((((((((((((-.--((..........)))---))))))))))))..........((((((....))))))...)))).......------------ ( -20.70, z-score = -3.31, R) >droYak2.chr2R 20145108 90 + 21139217 ---ACUUACAUCGUAAAUCAGUCAU-U--AGCUAUCAAUUCCUA---UGAUUGAUUUACACAUUGUUUUUGAUCGUUAUUGAUCACAUUGUAUUUGUCU------------ ---...((((..(((((((((((((-.--..............)---))))))))))))..........((((((....))))))...)))).......------------ ( -21.06, z-score = -3.72, R) >droEre2.scaffold_4845 19903002 90 + 22589142 ---UCUUACAUCGUAAAUCAGUCAU-U--AGCUAUCAAUUCCUA---UGAUUGAUUUACACAUUGGUUUUGAUUGUUAUUGAUCACAUUGCGUUCGUCU------------ ---........(((((....((((.-(--(((.(((((..((.(---((...........))).))..))))).)))).))))....))))).......------------ ( -18.20, z-score = -1.96, R) >droAna3.scaffold_12943 2073306 102 + 5039921 ---UCUUUCGUUGUAAAUCAGUCAU-U--AGCUAUCAAUUCCUU---UGAUUGAUUUACGCAUUGUUUUUGAUCGUUAUUGAUCAUAUUUUUUUUCACUCGUUAUAUAUAU ---......((.((((((((((((.-.--...............---))))))))))))))........((((((....)))))).......................... ( -18.73, z-score = -3.19, R) >dp4.chr4_group1 1218920 96 - 5278887 UCGUUUAUAUUCAUAAAUCAGUCAU-U--AGCUAUCAAUUGCUGGUUUGAUUGAUUUACUCAAUGCUUUUGAUCUUUUUGCCUGGUACCCUGGUUCUCU------------ .............((((((((((((-(--(((........)))))..))))))))))).((((.....)))).......(((.((...)).))).....------------ ( -19.00, z-score = -1.87, R) >droPer1.super_5 2825985 96 + 6813705 UCGUUUAUAUUCAUAAAUCAGUCAU-U--AGCUAUCAAUUGCUGGUUUGAUUGAUUUACUCAAUGCUUUUGAUCUUUUUGCCUGGUACCCUGGUUCUCU------------ .............((((((((((((-(--(((........)))))..))))))))))).((((.....)))).......(((.((...)).))).....------------ ( -19.00, z-score = -1.87, R) >droWil1.scaffold_181071 272945 83 - 1211509 -UUUUACGAAUCAUAAAUAAGUCACGA--AGCUAUCAAUUCUAU---UGAUUGAUUUAUAA---CCUUUUGAUCGUUAAUGAUCAUCCAUAA------------------- -...............((((((((...--....((((((...))---))))))))))))..---.....((((((....)))))).......------------------- ( -10.51, z-score = -0.47, R) >droVir3.scaffold_12963 5473621 94 - 20206255 ---GUUUGGCUUAUAAAUCCAUCAAGG--GGCUAUCAAUAUAUU--UUGAUUGAUUUAUAA---GGUUUAAAUCGUUGCUGAUCAUAUUUCUUAGUAGUUCAUU------- ---......((((((((((.(((((((--.............))--))))).)))))))))---)..........(((((((.........)))))))......------- ( -16.82, z-score = -0.99, R) >droMoj3.scaffold_6500 15220399 97 + 32352404 ---GUUUCGCUUGUAAAUCCAUCAGCGCAGCCUAUCAAUACAUU--UUGAUUGAUUUAAAG---UGCUUUAAUCGUCGCUGAUCAUAUUUCUUGCUGGUUUAUUU------ ---.....((..(.((((..((((((((.....(((((......--))))).((((.(((.---...)))))))).)))))))...)))))..))..........------ ( -16.60, z-score = -0.37, R) >droGri2.scaffold_14856 13692 95 - 51369 ---GUCUGCCUUGUAAAUCCUUCACUG--GGCUAUCGAUACGUU--UUGAUUGAUUUAUAA---GCUUUUAAUCAUUCAUGAUAAUAAUUCUGAGUAGGUGACUU------ ---(((.((.(((((((((..(((..(--(((.........)))--))))..)))))))))---)).....((((((((.((.......))))))).))))))..------ ( -17.50, z-score = -0.66, R) >consensus ___UCUUACAUCGUAAAUCAGUCAU_U__AGCUAUCAAUUCCUA___UGAUUGAUUUACACAUUGUUUUUGAUCGUUAUUGAUCAUAUUGUGUUCGUCU____________ ............(((((((.((((.......................)))).))))))).................................................... ( -6.08 = -6.15 + 0.07)

| Location | 21,838,988 – 21,839,082 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 65.61 |
| Shannon entropy | 0.70432 |
| G+C content | 0.32835 |
| Mean single sequence MFE | -19.85 |
| Consensus MFE | -6.08 |
| Energy contribution | -6.15 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.625267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 21838988 94 - 23011544 ---------AUGCGGAUUUACGGCACAGACAUGAGUCUUACA---UCGUAAAUCAGUCAU-U--AGCUAUCAAUUCCUA---UGAUUGAUUUACACAUUGUUUUUGAUCGUU ---------.....((((...((((.((((....))))....---..(((((((((((((-.--..............)---))))))))))))....))))...))))... ( -21.36, z-score = -1.97, R) >droSim1.chr2L 21412065 94 - 22036055 ---------AUGCGGAUUUACGGCACAGACAUGAGUCUUACA---UCGUAAAUCAGUCAU-C--AGCUAUCAAUUCCUA---UGAUUGAUUUACAAAUUGUUUUUGAUCGUU ---------.....((((...((((.((((....))))....---..(((((((((((((-.--((..........)))---))))))))))))....))))...))))... ( -21.30, z-score = -1.81, R) >droSec1.super_28 397605 94 - 873875 ---------AUGCGGAUUUACGGCACAGACAUGAGUCUUACA---UCGUAAAUCAGUCAU-C--AGCUAUCAAAUCCUA---UGAUUGAUUUACAAAUUGUUUUUGAUCGUU ---------.....((((...((((.((((....))))....---..(((((((((((((-.--((..........)))---))))))))))))....))))...))))... ( -21.30, z-score = -1.77, R) >droYak2.chr2R 20145130 94 + 21139217 ---------AGGCGGAUUUACGGCACAGAUAUGAAACUUACA---UCGUAAAUCAGUCAU-U--AGCUAUCAAUUCCUA---UGAUUGAUUUACACAUUGUUUUUGAUCGUU ---------.(.((......)).)...(((..(((((.....---..(((((((((((((-.--..............)---)))))))))))).....)))))..)))... ( -20.76, z-score = -2.20, R) >droEre2.scaffold_4845 19903024 94 + 22589142 ---------AGGCGGAUUUACGGCACAGACAUGAGUCUUACA---UCGUAAAUCAGUCAU-U--AGCUAUCAAUUCCUA---UGAUUGAUUUACACAUUGGUUUUGAUUGUU ---------.(((.(((((((((...((((....))))....---))))))))).)))..-.--(((.(((((..((.(---((...........))).))..))))).))) ( -24.80, z-score = -2.98, R) >droAna3.scaffold_12943 2073340 97 + 5039921 ------AUUGGGGGGAUUUACGGCCCAGACAUGAGUCUUUCG---UUGUAAAUCAGUCAU-U--AGCUAUCAAUUCCUU---UGAUUGAUUUACGCAUUGUUUUUGAUCGUU ------....(..(((((((.(.......).)))))))..)(---(.((((((((((((.-.--...............---))))))))))))))................ ( -22.93, z-score = -1.19, R) >dp4.chr4_group1 1218942 99 - 5278887 ----------UUUGGAUUUAUGAUUAAAACAUGACUCGUUUAUAUUCAUAAAUCAGUCAU-U--AGCUAUCAAUUGCUGGUUUGAUUGAUUUACUCAAUGCUUUUGAUCUUU ----------...........(((((((((((((...(........).((((((((((((-(--(((........)))))..))))))))))).)).))).))))))))... ( -20.50, z-score = -2.14, R) >droPer1.super_5 2826007 99 + 6813705 ----------UUUGGAUUUAUGAUUAAAACAUGACUCGUUUAUAUUCAUAAAUCAGUCAU-U--AGCUAUCAAUUGCUGGUUUGAUUGAUUUACUCAAUGCUUUUGAUCUUU ----------...........(((((((((((((...(........).((((((((((((-(--(((........)))))..))))))))))).)).))).))))))))... ( -20.50, z-score = -2.14, R) >droWil1.scaffold_181071 272960 102 - 1211509 AUAUAUUUGUUUUAGAAACACUCCAU-GACAUGGCUUUUACGAA-UCAUAAAUAAGUCACGA--AGCUAUCAAUUCUAU---UGAUUGAUUUAUAAC---CUUUUGAUCGUU .......((((...((.....))...-))))........((((.-(((.((((((((((...--....((((((...))---))))))))))))...---.)).))))))). ( -10.21, z-score = 1.13, R) >droVir3.scaffold_12963 5473648 92 - 20206255 -------UAACGAAAUAAUAUGACCCAUAGUU------GUUUGGCUUAUAAAUCCAUCAAGG--GGCUAUCAAUAUAUU--UUGAUUGAUUUAUAAG---GUUUAAAUCGUU -------.(((((...........(((.....------...)))((((((((((.(((((((--.............))--))))).))))))))))---.......))))) ( -19.32, z-score = -1.68, R) >droMoj3.scaffold_6500 15220427 93 + 32352404 -------GGUUGAUAUUAUAAGACCUCUA-UU------GUUUCGCUUGUAAAUCCAUCAGCGCAGCCUAUCAAUACAUU--UUGAUUGAUUUAAAGU---GCUUUAAUCGUC -------(((.(((.(((((((....(..-..------).....)))))))))).)))(((((.....((((((.....--...)))))).....))---)))......... ( -16.40, z-score = -0.96, R) >droGri2.scaffold_14856 13720 98 - 51369 -------UAUGGAAUGAAUAAGCACCUUAGACAUGUAGGUCUGCCUUGUAAAUCCUUCACUG--GGCUAUCGAUACGUU--UUGAUUGAUUUAUAAG---CUUUUAAUCAUU -------.....(((((..((((....(((((......)))))..(((((((((..(((..(--(((.........)))--))))..))))))))))---)))....))))) ( -18.80, z-score = -0.48, R) >consensus _________AUGCGGAUUUACGGCACAGACAUGAGUCUUACA___UCGUAAAUCAGUCAU_U__AGCUAUCAAUUCCUA___UGAUUGAUUUACACAUUGUUUUUGAUCGUU ...............................................(((((((.((((.......................)))).))))))).................. ( -6.08 = -6.15 + 0.07)
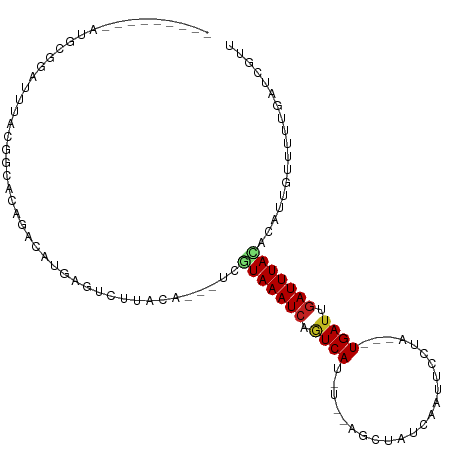
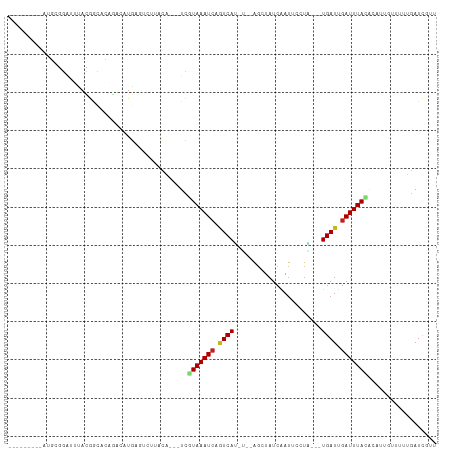
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:57:11 2011