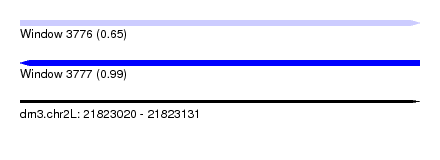
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,823,020 – 21,823,131 |
| Length | 111 |
| Max. P | 0.994230 |

| Location | 21,823,020 – 21,823,131 |
|---|---|
| Length | 111 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.00 |
| Shannon entropy | 0.14788 |
| G+C content | 0.40178 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -21.35 |
| Energy contribution | -21.43 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.651234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
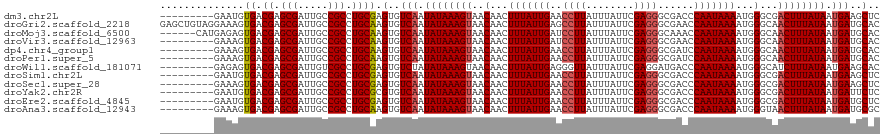
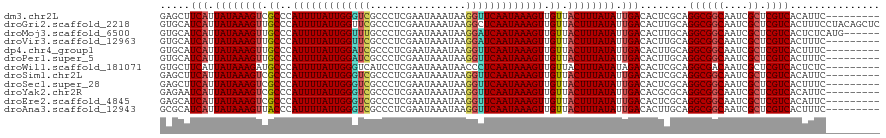
>dm3.chr2L 21823020 111 + 23011544 ---------GAAUGUGACGAGCGAUUGCCGCCUGCGAGUGUCAAUAUAAAGUAACAACUUUAUUGAACCUUAUUUAUUCGAGGGCGACCCAAUAAAAUGGGCGACUUUAUAAUGAAGCUC ---------.........((((......(((((.((((((((((((..((((....))))))))))........)))))).))))).((((......))))...............)))) ( -27.70, z-score = -2.02, R) >droGri2.scaffold_2218 2630 120 + 3229 GAGCUGUAGGAAAGUGACGAGCGAUUGCCGCCUGCAAGUGUCAAUAUAAAGUAACAACUUUAUUGAGCCUUAUUUAUUCGAGGGCGAACCAAUAAAAUGGGCAACUUUAUAAUGAUGCAC ....((((((...(..((....).)..)..)))))).((((((.((((((((..(...(((((((.(((((.........)))))....)))))))...)...)))))))).)))))).. ( -32.10, z-score = -2.18, R) >droMoj3.scaffold_6500 17107430 114 + 32352404 ------CAUGAGAGUGACGAGCGAUUGCCGCCUGCAAGUGUCAAUAUAAAGUAACAACUUUAUUGAUCCUUAUUUAUUCGAGGGCAAACCAAUAAAAUGGGCAACUUUAUAAUGAUGCAC ------(((....))).((.(((.....))).))...((((((.((((((((..........(((.(((((........)))))))).(((......)))...)))))))).)))))).. ( -26.50, z-score = -1.61, R) >droVir3.scaffold_12963 14708674 111 - 20206255 ---------GAAAGUGACGAGCGAUUGCCGCCUGCAAGUGUCAAUAUAAAGUAACAACUUUAUUGAUCCUUAUUUAUUCGAGGGCGAACCAAUAAAAUGGGCAACUUUAUAAUGAUGCAC ---------.....((.((.(((.....))).)))).((((((.((((((((..........(((.(((((........)))))))).(((......)))...)))))))).)))))).. ( -25.20, z-score = -1.49, R) >dp4.chr4_group1 1200294 111 + 5278887 ---------GAAAGUGACGAGCGAUUGCCGCCUGCAAGUGUCAAUAUAAAGUAACAACUUUAUUGAACCUUAUUUAUUCGAGGGCGAUCCAAUAAAAUGGGCAACUUUAUAAUGAUGCAC ---------.....((.((.(((.....))).)))).((((((.((((((((..(...(((((((..((((........))))......)))))))...)...)))))))).)))))).. ( -24.20, z-score = -1.16, R) >droPer1.super_5 2807415 111 - 6813705 ---------GAAAGUGACGAGCGAUUGCCGCCUGCAAGUGUCAAUAUAAAGUAACAACUUUAUUGAACCUUAUUUAUUCGAGGGCGAUCCAAUAAAAUGGGCAACUUUAUAAUGAUGCAC ---------.....((.((.(((.....))).)))).((((((.((((((((..(...(((((((..((((........))))......)))))))...)...)))))))).)))))).. ( -24.20, z-score = -1.16, R) >droWil1.scaffold_181071 255483 111 + 1211509 ---------GAGAGUGACGAGCGAUUGUCGCCUGCGAGUGUCUAUAUAAAGUAACAACUUUAUUGAGGGUUAUUUAUUCGAGGAUGACCCAAUAAAAUGGGCAUCUUUAUAAUGAAGCAC ---------.((.(((((((....)))))))))((((.(.((...(((((((....))))))).)).).)).((((((.(((((((.((((......)))))))))))..)))))))).. ( -32.60, z-score = -3.37, R) >droSim1.chr2L 21397829 111 + 22036055 ---------GAAUGUGACGAGCGAUUGCCGCCUGCGAGUGUCAAUAUAAAGUAACAACUUUAUUGAACCUUAUUUAUUCGAGGGCGACCCAAUAAAAUGGGCGACUUUAUAAUGAAGCUC ---------.........((((......(((((.((((((((((((..((((....))))))))))........)))))).))))).((((......))))...............)))) ( -27.70, z-score = -2.02, R) >droSec1.super_28 383540 111 + 873875 ---------GAAAGUGACGAGCGAUUGCCGCCUGCGAGUGUCAAUAUAAAGUAACAACUUUAUUGAACCUUAUUUAUUCGAGGGCGACCCAAUAAAAUGGGCGACUUUAUAAUGAAGCUC ---------.........((((......(((((.((((((((((((..((((....))))))))))........)))))).))))).((((......))))...............)))) ( -27.70, z-score = -2.03, R) >droYak2.chr2R 20127403 111 - 21139217 ---------GAAUGUGACGAGCGAUUGCCGCCUGCGCGUGUCAAUAUAAAGUAACAACUUUAUUGAACCUUAUUUAUUCGAGGGCGACCCAAUAAAAUGGGCGACUUUAUAAUGAUUCUC ---------..(((((.((.(((.....))).)))))))((((.((((((((...............((((........)))).((.((((......)))))))))))))).)))).... ( -28.70, z-score = -2.14, R) >droEre2.scaffold_4845 19886125 111 - 22589142 ---------GAAUGUGACGAGCGAUUGCCGCCUGCGAGUGUCAAUAUAAAGUAACAACUUUAUUGAACCUUAUUUAUUCGAGGGCGACCCAAUAAAAUGGGCGACUUUAUAAUGAUGCUC ---------......(.((.(((.....))).)))((((((((.((((((((...............((((........)))).((.((((......)))))))))))))).)))))))) ( -30.40, z-score = -2.89, R) >droAna3.scaffold_12943 2059233 111 - 5039921 ---------GAAAGUGACGAGCGAUUGCCGCCUGCAAGUGUCAAUAUAAAGUAACAACUUUAUUGAACCUUAUUUAUUCGAGGGCGACCCAAUAAAAUGGGUAACUUUAUAAUGAUGCGC ---------.....((.((.(((.....))).)))).((((((.((((((((...............((((........))))...(((((......))))).)))))))).)))))).. ( -27.80, z-score = -2.20, R) >consensus _________GAAAGUGACGAGCGAUUGCCGCCUGCAAGUGUCAAUAUAAAGUAACAACUUUAUUGAACCUUAUUUAUUCGAGGGCGACCCAAUAAAAUGGGCAACUUUAUAAUGAUGCAC ..............((.((.(((.....))).)))).(..(((.((((((((..(...(((((((..((((........))))......)))))))...)...)))))))).)))..).. (-21.35 = -21.43 + 0.08)
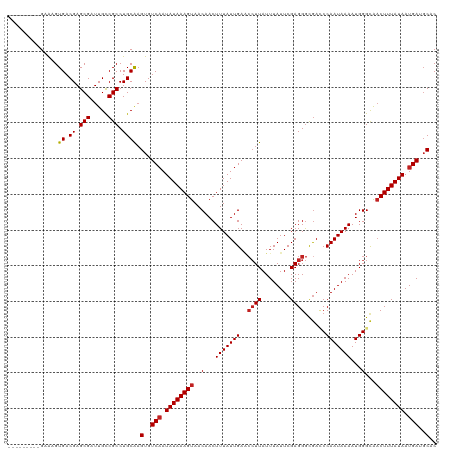
| Location | 21,823,020 – 21,823,131 |
|---|---|
| Length | 111 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.00 |
| Shannon entropy | 0.14788 |
| G+C content | 0.40178 |
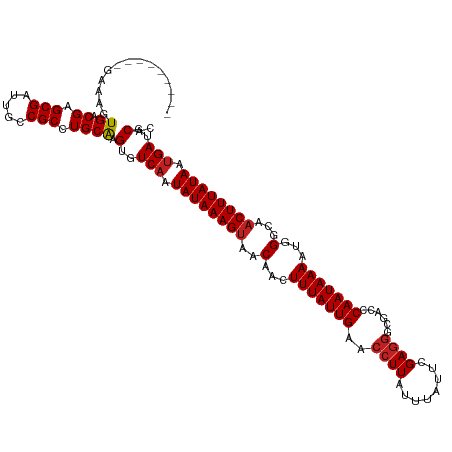
| Mean single sequence MFE | -29.96 |
| Consensus MFE | -24.88 |
| Energy contribution | -24.83 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.994230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
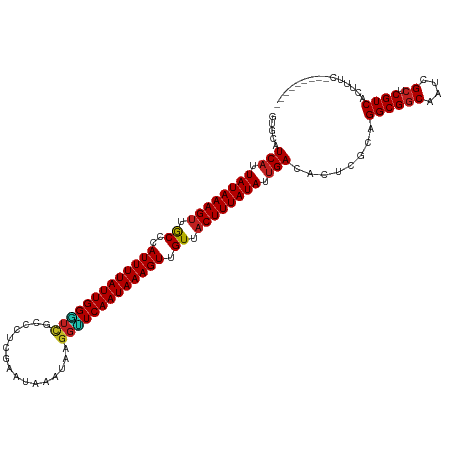
>dm3.chr2L 21823020 111 - 23011544 GAGCUUCAUUAUAAAGUCGCCCAUUUUAUUGGGUCGCCCUCGAAUAAAUAAGGUUCAAUAAAGUUGUUACUUUAUAUUGACACUCGCAGGCGGCAAUCGCUCGUCACAUUC--------- (((..(((.((((((((.((..(((((((((..((................))..))))))))).)).)))))))).)))..)))...((((((....)).))))......--------- ( -31.59, z-score = -3.88, R) >droGri2.scaffold_2218 2630 120 - 3229 GUGCAUCAUUAUAAAGUUGCCCAUUUUAUUGGUUCGCCCUCGAAUAAAUAAGGCUCAAUAAAGUUGUUACUUUAUAUUGACACUUGCAGGCGGCAAUCGCUCGUCACUUUCCUACAGCUC .(((((((.((((((((.((..((((((((((...(((.............))))))))))))).)).)))))))).)))....))))((((((....)).))))............... ( -29.42, z-score = -2.41, R) >droMoj3.scaffold_6500 17107430 114 - 32352404 GUGCAUCAUUAUAAAGUUGCCCAUUUUAUUGGUUUGCCCUCGAAUAAAUAAGGAUCAAUAAAGUUGUUACUUUAUAUUGACACUUGCAGGCGGCAAUCGCUCGUCACUCUCAUG------ .(((((((.((((((((.((..(((((((((((((................))))))))))))).)).)))))))).)))....))))((((((....)).)))).........------ ( -29.19, z-score = -2.68, R) >droVir3.scaffold_12963 14708674 111 + 20206255 GUGCAUCAUUAUAAAGUUGCCCAUUUUAUUGGUUCGCCCUCGAAUAAAUAAGGAUCAAUAAAGUUGUUACUUUAUAUUGACACUUGCAGGCGGCAAUCGCUCGUCACUUUC--------- .(((((((.((((((((.((..(((((((((((((................))))))))))))).)).)))))))).)))....))))((((((....)).))))......--------- ( -31.59, z-score = -3.86, R) >dp4.chr4_group1 1200294 111 - 5278887 GUGCAUCAUUAUAAAGUUGCCCAUUUUAUUGGAUCGCCCUCGAAUAAAUAAGGUUCAAUAAAGUUGUUACUUUAUAUUGACACUUGCAGGCGGCAAUCGCUCGUCACUUUC--------- .(((((((.((((((((.((..(((((((((((((................))))))))))))).)).)))))))).)))....))))((((((....)).))))......--------- ( -31.59, z-score = -3.86, R) >droPer1.super_5 2807415 111 + 6813705 GUGCAUCAUUAUAAAGUUGCCCAUUUUAUUGGAUCGCCCUCGAAUAAAUAAGGUUCAAUAAAGUUGUUACUUUAUAUUGACACUUGCAGGCGGCAAUCGCUCGUCACUUUC--------- .(((((((.((((((((.((..(((((((((((((................))))))))))))).)).)))))))).)))....))))((((((....)).))))......--------- ( -31.59, z-score = -3.86, R) >droWil1.scaffold_181071 255483 111 - 1211509 GUGCUUCAUUAUAAAGAUGCCCAUUUUAUUGGGUCAUCCUCGAAUAAAUAACCCUCAAUAAAGUUGUUACUUUAUAUAGACACUCGCAGGCGACAAUCGCUCGUCACUCUC--------- (((((....((((((((((((((......)))).))))........((((((..........))))))..)))))).)).)))..((..(((.....)))..)).......--------- ( -23.00, z-score = -2.33, R) >droSim1.chr2L 21397829 111 - 22036055 GAGCUUCAUUAUAAAGUCGCCCAUUUUAUUGGGUCGCCCUCGAAUAAAUAAGGUUCAAUAAAGUUGUUACUUUAUAUUGACACUCGCAGGCGGCAAUCGCUCGUCACAUUC--------- (((..(((.((((((((.((..(((((((((..((................))..))))))))).)).)))))))).)))..)))...((((((....)).))))......--------- ( -31.59, z-score = -3.88, R) >droSec1.super_28 383540 111 - 873875 GAGCUUCAUUAUAAAGUCGCCCAUUUUAUUGGGUCGCCCUCGAAUAAAUAAGGUUCAAUAAAGUUGUUACUUUAUAUUGACACUCGCAGGCGGCAAUCGCUCGUCACUUUC--------- (((..(((.((((((((.((..(((((((((..((................))..))))))))).)).)))))))).)))..)))...((((((....)).))))......--------- ( -31.59, z-score = -3.80, R) >droYak2.chr2R 20127403 111 + 21139217 GAGAAUCAUUAUAAAGUCGCCCAUUUUAUUGGGUCGCCCUCGAAUAAAUAAGGUUCAAUAAAGUUGUUACUUUAUAUUGACACGCGCAGGCGGCAAUCGCUCGUCACAUUC--------- .....(((.((((((((.((..(((((((((..((................))..))))))))).)).)))))))).)))........((((((....)).))))......--------- ( -27.29, z-score = -2.01, R) >droEre2.scaffold_4845 19886125 111 + 22589142 GAGCAUCAUUAUAAAGUCGCCCAUUUUAUUGGGUCGCCCUCGAAUAAAUAAGGUUCAAUAAAGUUGUUACUUUAUAUUGACACUCGCAGGCGGCAAUCGCUCGUCACAUUC--------- (((..(((.((((((((.((..(((((((((..((................))..))))))))).)).)))))))).)))..)))...((((((....)).))))......--------- ( -31.59, z-score = -3.97, R) >droAna3.scaffold_12943 2059233 111 + 5039921 GCGCAUCAUUAUAAAGUUACCCAUUUUAUUGGGUCGCCCUCGAAUAAAUAAGGUUCAAUAAAGUUGUUACUUUAUAUUGACACUUGCAGGCGGCAAUCGCUCGUCACUUUC--------- ..((((((.((((((((.((..(((((((((..((................))..))))))))).)).)))))))).)))....))).((((((....)).))))......--------- ( -29.49, z-score = -3.35, R) >consensus GUGCAUCAUUAUAAAGUUGCCCAUUUUAUUGGGUCGCCCUCGAAUAAAUAAGGUUCAAUAAAGUUGUUACUUUAUAUUGACACUCGCAGGCGGCAAUCGCUCGUCACUUUC_________ .....(((.((((((((.((..(((((((((((((................))))))))))))).)).)))))))).)))........((((((....)).))))............... (-24.88 = -24.83 + -0.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:57:08 2011