| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,801,727 – 21,801,821 |
| Length | 94 |
| Max. P | 0.948743 |

| Location | 21,801,727 – 21,801,821 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 78.81 |
| Shannon entropy | 0.45021 |
| G+C content | 0.43210 |
| Mean single sequence MFE | -18.67 |
| Consensus MFE | -10.42 |
| Energy contribution | -11.09 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
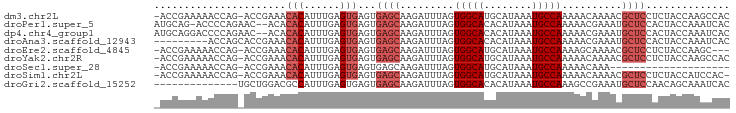
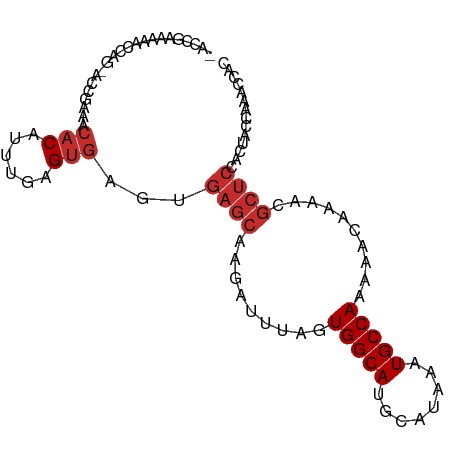
>dm3.chr2L 21801727 94 + 23011544 -ACCGAAAAACCAG-ACCGAAACACAUUUGAGUGAGUGAGCAAGAUUUAGUGGCAUGCAUAAAUGCCAAAAACAAAACGCUCCUCUACCAAGCCAC -.............-...........((((.(((((.((((.........((((((......))))))..........))))))).)))))).... ( -20.51, z-score = -2.86, R) >droPer1.super_5 2778558 93 - 6813705 AUGCAG-ACCCCAGAAC--ACACACAUUUGAGUGAGUGAGCAAGAUUUAGUGGCACACAUAAAUGCCAAAAACGAAAUGCUCCACUACCAAAUCAC ......-..........--......(((((.((.((((((((........(((((........))))).........)))).)))))))))))... ( -18.23, z-score = -1.81, R) >dp4.chr4_group1 1171688 94 + 5278887 AUGCAGGACCCCAGAAC--ACACACAUUUGAGUGAGUGAGCAAGAUUUAGUGGCACACAUAAAUGCCAAAAACGAAAUGCUCCACUACCAAAUCAC .....((.........(--((.((....)).)))((((((((........(((((........))))).........)))).)))).))....... ( -19.13, z-score = -1.55, R) >droAna3.scaffold_12943 2036648 87 - 5039921 ---------ACCAGCACCGAAACACAUUUGAGUGAGUGAGCAAGAUUUAGUGGCACACAUAAAUGCCAAAAACGAAAUGCUCCACUACCAAAUCAC ---------................(((((.((.((((((((........(((((........))))).........)))).)))))))))))... ( -18.23, z-score = -2.29, R) >droEre2.scaffold_4845 19861522 91 - 22589142 -ACCGAAAAACCAG-ACCGAAACACAUUUGAGUGAGUGAGCAAGAUUUAGUGGCAUGCAUAAAUGCCAAAAGCAAAACGCUCCUCUACCAAGC--- -.............-...........((((.(((((.((((.........((((((......))))))..........))))))).)))))).--- ( -20.51, z-score = -2.46, R) >droYak2.chr2R 20104790 94 - 21139217 -ACCGAAAAACCAG-ACCGAAACACAUUUGAGUGAGUGAGCAAGAUUUAGUGGCAUGCAUAAAUGCCAAAAACAAAACGCUCCUCUACCAAGCCAC -.............-...........((((.(((((.((((.........((((((......))))))..........))))))).)))))).... ( -20.51, z-score = -2.86, R) >droSec1.super_28 363629 74 + 873875 -ACCGAAAAACCAG-ACCGAAACACAUUUGAGUGAGUGAGCAAGAUUUAGUGGCAUGCAUAAAUGCCAAAAACAAA-------------------- -.............-.........(((((....)))))............((((((......))))))........-------------------- ( -10.60, z-score = -0.75, R) >droSim1.chr2L 21376827 93 + 22036055 -ACCGAAAAACCAG-ACCGAAACACAUUUGAGUGAGUGAGCAAGAUUUAGUGGCAUGCAUAAAUGCCAAAAACAAAACGCUCCUCUACCAUCCAC- -.............-.............((.(((((.((((.........((((((......))))))..........))))))).)))).....- ( -19.01, z-score = -2.53, R) >droGri2.scaffold_15252 9053341 82 - 17193109 --------------UGCUGGACGCCAUUUGAGUGAGUGAGCAAGAUUUAGUGGCACACAUAAAUGCCAAAGCCGAAAUGCUCCAACAGCAAAUCAC --------------(((((..((((....).))).(.(((((........(((((........))))).........))))))..)))))...... ( -21.33, z-score = -1.37, R) >consensus _ACCGAAAAACCAG_ACCGAAACACAUUUGAGUGAGUGAGCAAGAUUUAGUGGCAUGCAUAAAUGCCAAAAACAAAACGCUCCACUACCAAACCAC ......................(((......)))...((((.........(((((........)))))..........)))).............. (-10.42 = -11.09 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:57:04 2011