| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,783,703 – 21,783,810 |
| Length | 107 |
| Max. P | 0.971032 |

| Location | 21,783,703 – 21,783,804 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 60.88 |
| Shannon entropy | 0.78232 |
| G+C content | 0.45963 |
| Mean single sequence MFE | -28.09 |
| Consensus MFE | -9.52 |
| Energy contribution | -10.25 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.971032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
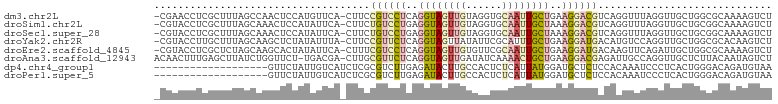
>dm3.chr2L 21783703 101 + 23011544 -CGAACCUCGCUUUAGCCAACUCCAUGUUCA-CUUCCGUCCUCAGGUAGUUGUAGGUGCAAUUGCUGAAGGACGUCAGGUUUAGGUUGCUGGCGCAAAAGUCU -........(((((.((((.(.((.((..(.-((..((((((..(((((((((....)))))))))..))))))..)))..))))..).))))...))))).. ( -32.90, z-score = -1.87, R) >droSim1.chr2L 21357372 101 + 22036055 -CGUACCUCGCUUUAGCAAACUCCAUAUUCA-CUUCUGUCCUGAGGUAGUUGUAGGUGCAAUUGCUAAAGGACGUCAGGUUUAGGUUGCUGCGGCAAAAGUCU -........(((.((((((...((.((..(.-((..((((((..(((((((((....)))))))))..))))))..)))..)))))))))).)))........ ( -31.30, z-score = -2.00, R) >droSec1.super_28 343741 101 + 873875 -CGUACCUCGCUUUAGCAAACUCCAUAUUCA-CUUCUGUCCUGAGGUAGUUGUAGGUGCAAUUGCUAAAGGACGUCAGGUUUAGGUUGCUGCGGCAAAAGUCU -........(((.((((((...((.((..(.-((..((((((..(((((((((....)))))))))..))))))..)))..)))))))))).)))........ ( -31.30, z-score = -2.00, R) >droYak2.chr2R 20081945 101 - 21139217 -CGUACCUUGCUUUAGCAAGCUCUAUAUUUA-CUUCCGUUCUCAGGUAGUUAUAUUCGCAUUUGCUGAAGGAUGACAUGUCCAGGUUGCUGGCGCACAAGUCU -.........(((((((((((...((((..(-((.((.......)).))).))))..))..)))))))))...(((.(((((((....))))...))).))). ( -25.30, z-score = -0.56, R) >droEre2.scaffold_4845 19843636 101 - 22589142 -CGUACCUCGCUCUAGCAAGCACUAUAUUCA-CUUUCGUCCUCAGGUAGUUGUGUUCGCAAUUGCUGAAGGAUGACAAGUUCAGAUUGCUGGCGCAAAAGUCU -........((.(((((((...((......(-((((((((((..(((((((((....)))))))))..))))))).))))..)).))))))).))........ ( -33.80, z-score = -2.97, R) >droAna3.scaffold_12943 2021146 101 - 5039921 ACAACUUUGAGCUUAUCUGGUUCU-UGACGA-CUUGCGUUCUCAGGUAGUUGAUAUCAAAACUGCUGAAGGACGAGAUUGCCAGGUUGCUCUUACAAUAGUCU ........((((..(((((((..(-(.....-....((((((..(((((((........)))))))..)))))).))..))))))).))))............ ( -30.50, z-score = -2.11, R) >dp4.chr4_group1 1151706 84 + 5278887 -------------------GUUCUAUUGUCAUCUCGCGUCUUGAGAUACUUGCCACUCUCAUUAUGGAUGCUCUCCACAAAUCCCUCACUGGGACAGAUGUAA -------------------..........(((((.(((((((((((..........)))))....))))))..........((((.....)))).)))))... ( -19.80, z-score = -1.58, R) >droPer1.super_5 2757695 84 - 6813705 -------------------GUUCUAUUGUCAUCUCGCGUCUUGAGAUACUUGCCACUCUCAUUAUGGAUGCUCUCCACAAAUCCCUCACUGGGACAGAUGUAA -------------------..........(((((.(((((((((((..........)))))....))))))..........((((.....)))).)))))... ( -19.80, z-score = -1.58, R) >consensus _CGUACCUCGCUUUAGCAAGCUCUAUAUUCA_CUUCCGUCCUCAGGUAGUUGUAAGUGCAAUUGCUGAAGGACGACAGGUUUAGGUUGCUGGGGCAAAAGUCU ....................................((((((..((((((((......))))))))..))))))............................. ( -9.52 = -10.25 + 0.73)

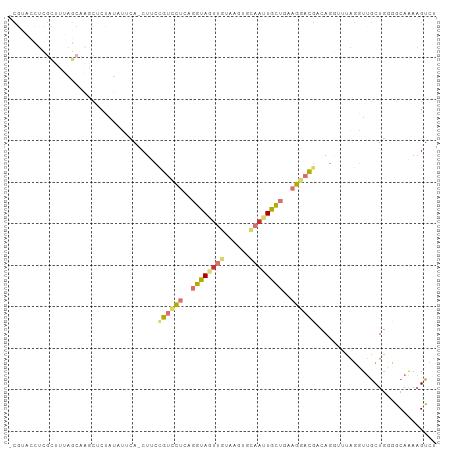
| Location | 21,783,709 – 21,783,810 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 62.24 |
| Shannon entropy | 0.75757 |
| G+C content | 0.45532 |
| Mean single sequence MFE | -28.38 |
| Consensus MFE | -9.52 |
| Energy contribution | -10.25 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
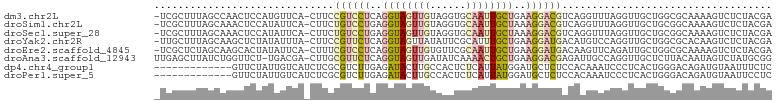
>dm3.chr2L 21783709 101 + 23011544 -UCGCUUUAGCCAACUCCAUGUUCA-CUUCCGUCCUCAGGUAGUUGUAGGUGCAAUUGCUGAAGGACGUCAGGUUUAGGUUGCUGGCGCAAAAGUCUCUACGA -..(((((.((((.(.((.((..(.-((..((((((..(((((((((....)))))))))..))))))..)))..))))..).))))...)))))........ ( -32.90, z-score = -1.92, R) >droSim1.chr2L 21357378 101 + 22036055 -UCGCUUUAGCAAACUCCAUAUUCA-CUUCUGUCCUGAGGUAGUUGUAGGUGCAAUUGCUAAAGGACGUCAGGUUUAGGUUGCUGCGGCAAAAGUCUCUACGA -..(((.((((((...((.((..(.-((..((((((..(((((((((....)))))))))..))))))..)))..)))))))))).))).............. ( -31.30, z-score = -1.91, R) >droSec1.super_28 343747 101 + 873875 -UCGCUUUAGCAAACUCCAUAUUCA-CUUCUGUCCUGAGGUAGUUGUAGGUGCAAUUGCUAAAGGACGUCAGGUUUAGGUUGCUGCGGCAAAAGUCUCUACGA -..(((.((((((...((.((..(.-((..((((((..(((((((((....)))))))))..))))))..)))..)))))))))).))).............. ( -31.30, z-score = -1.91, R) >droYak2.chr2R 20081951 101 - 21139217 -UUGCUUUAGCAAGCUCUAUAUUUA-CUUCCGUUCUCAGGUAGUUAUAUUCGCAUUUGCUGAAGGAUGACAUGUCCAGGUUGCUGGCGCACAAGUCUCUACGA -...(((((((((((...((((..(-((.((.......)).))).))))..))..)))))))))((.(((.(((((((....))))...))).)))))..... ( -26.00, z-score = -0.76, R) >droEre2.scaffold_4845 19843642 101 - 22589142 -UCGCUCUAGCAAGCACUAUAUUCA-CUUUCGUCCUCAGGUAGUUGUGUUCGCAAUUGCUGAAGGAUGACAAGUUCAGAUUGCUGGCGCAAAAGUCUCUACGA -..((.(((((((...((......(-((((((((((..(((((((((....)))))))))..))))))).))))..)).))))))).)).............. ( -33.80, z-score = -2.73, R) >droAna3.scaffold_12943 2021152 101 - 5039921 UUGAGCUUAUCUGGUUCU-UGACGA-CUUGCGUUCUCAGGUAGUUGAUAUCAAAACUGCUGAAGGACGAGAUUGCCAGGUUGCUCUUACAAUAGUCUAUGCGG ..((((..(((((((..(-(.....-....((((((..(((((((........)))))))..)))))).))..))))))).)))).................. ( -30.50, z-score = -1.74, R) >dp4.chr4_group1 1151706 90 + 5278887 -------------GUUCUAUUGUCAUCUCGCGUCUUGAGAUACUUGCCACUCUCAUUAUGGAUGCUCUCCACAAAUCCCUCACUGGGACAGAUGUAAUUUCUC -------------.....((((.(((((.(((((((((((..........)))))....))))))..........((((.....)))).)))))))))..... ( -20.60, z-score = -1.90, R) >droPer1.super_5 2757695 90 - 6813705 -------------GUUCUAUUGUCAUCUCGCGUCUUGAGAUACUUGCCACUCUCAUUAUGGAUGCUCUCCACAAAUCCCUCACUGGGACAGAUGUAAUUCCUC -------------.....((((.(((((.(((((((((((..........)))))....))))))..........((((.....)))).)))))))))..... ( -20.60, z-score = -1.81, R) >consensus _UCGCUUUAGCAAGCUCUAUAUUCA_CUUCCGUCCUCAGGUAGUUGUAAGUGCAAUUGCUGAAGGACGACAGGUUUAGGUUGCUGGGGCAAAAGUCUCUACGA ..............................((((((..((((((((......))))))))..))))))................................... ( -9.52 = -10.25 + 0.73)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:57:02 2011