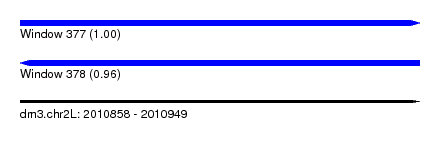
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,010,858 – 2,010,949 |
| Length | 91 |
| Max. P | 0.996402 |

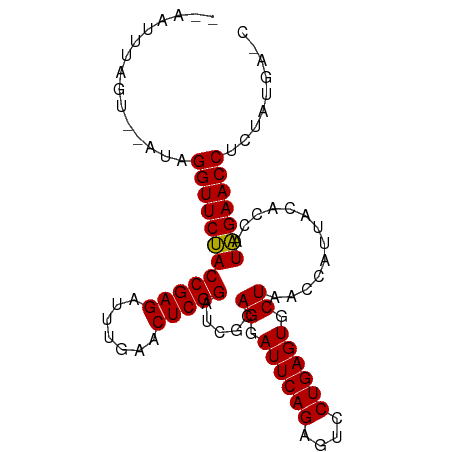
| Location | 2,010,858 – 2,010,949 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 85.78 |
| Shannon entropy | 0.31399 |
| G+C content | 0.45894 |
| Mean single sequence MFE | -27.27 |
| Consensus MFE | -25.71 |
| Energy contribution | -25.25 |
| Covariance contribution | -0.46 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.996402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2010858 91 + 23011544 --AAUGUAGU--AUAGGUUCUACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUAGAACCUCUAUGGGC --..((((((--...((((...(((((.......))))).....((.((((((....)))))).))))))))))))((((((.....)))))).. ( -29.10, z-score = -2.63, R) >droGri2.scaffold_15252 165606 80 + 17193109 ----AUUUGA--CUAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACC--------- ----......--...((((((((((((.......))))).....((.((((((....)))))).)).............)))))))--------- ( -25.20, z-score = -2.87, R) >droMoj3.scaffold_6500 29330957 93 + 32352404 AAGGUCACACCGACAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCACUGCGU-- ...(((.....))).((((((((((((.......))))).....((.((((((....)))))).)).............))))))).......-- ( -29.10, z-score = -2.08, R) >droVir3.scaffold_12963 7607094 91 - 20206255 --ACUUUAAUAAUCAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCGUUGAGA-- --.............((((((((((((.......))))).....((.((((((....)))))).)).............))))))).......-- ( -26.90, z-score = -2.50, R) >droWil1.scaffold_180708 11651640 91 - 12563649 --AUCUUAAC--UCAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCGUUGAAGAC --.((((...--...((((((((((((.......))))).....((.((((((....)))))).)).............)))))))....)))). ( -28.70, z-score = -2.82, R) >droPer1.super_1 10242609 91 + 10282868 --AUUUCACU--UCAGGUUCUACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUAGAACCGCUUUGAAC --..((((..--...((((((((((((.......))))).....((.((((((....)))))).)).............)))))))....)))). ( -26.10, z-score = -2.90, R) >dp4.chr4_group3 8790860 91 + 11692001 --AUUUCACU--UCAGGUUCUACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUAGAACCGCUUUGAAC --..((((..--...((((((((((((.......))))).....((.((((((....)))))).)).............)))))))....)))). ( -26.10, z-score = -2.90, R) >droEre2.scaffold_4929 2056848 91 + 26641161 --AAGUCCGU--AUAGGUUCUACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUAGAACCUCUAUGAGC --..(..(((--(.(((((((((((((.......))))).....((.((((((....)))))).)).............)))))))).))))..) ( -29.70, z-score = -3.35, R) >droYak2.chr2L 1995823 91 + 22324452 --AACUUAGU--AUAGGUUCUACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUAGAACCUCUAUGAGC --..((((..--..(((((((((((((.......))))).....((.((((((....)))))).)).............))))))))...)))). ( -27.40, z-score = -2.93, R) >droSec1.super_14 1957687 91 + 2068291 --AAUUUAGU--CUAGGUUCUACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUAGAACCUCUAUGGGU --........--...((((...(((((.......))))).....((.((((((....)))))).))))))......((((((.....)))))).. ( -26.80, z-score = -1.87, R) >droSim1.chr2L 1980529 86 + 22036055 --AAUGUAGU--CUAGGUUCUACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUAGAACCUCUA----- --........--..(((((((((((((.......))))).....((.((((((....)))))).)).............))))))))...----- ( -24.90, z-score = -2.10, R) >consensus __AAUUUAGU__AUAGGUUCUACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUAGAACCUCUAUGA_C ...............((((((((((((.......))))).....((.((((((....)))))).)).............)))))))......... (-25.71 = -25.25 + -0.46)

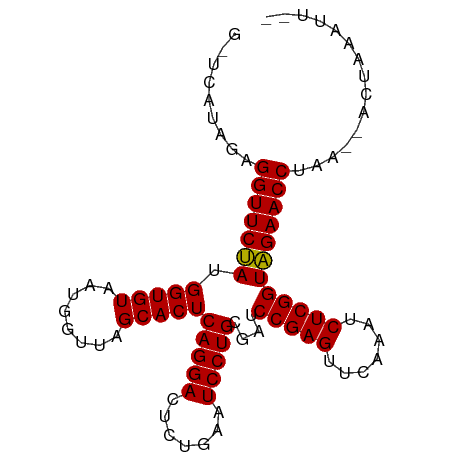
| Location | 2,010,858 – 2,010,949 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 85.78 |
| Shannon entropy | 0.31399 |
| G+C content | 0.45894 |
| Mean single sequence MFE | -25.97 |
| Consensus MFE | -24.79 |
| Energy contribution | -24.33 |
| Covariance contribution | -0.46 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.956025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2010858 91 - 23011544 GCCCAUAGAGGUUCUAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUAGAACCUAU--ACUACAUU-- ........((((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))))..--........-- ( -24.90, z-score = -1.34, R) >droGri2.scaffold_15252 165606 80 - 17193109 ---------GGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCUAG--UCAAAU---- ---------(((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))...--......---- ( -25.40, z-score = -2.22, R) >droMoj3.scaffold_6500 29330957 93 - 32352404 --ACGCAGUGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCUGUCGGUGUGACCUU --((((...(((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))).....))))...... ( -29.00, z-score = -0.74, R) >droVir3.scaffold_12963 7607094 91 + 20206255 --UCUCAACGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCUGAUUAUUAAAGU-- --.......(((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))).............-- ( -25.70, z-score = -1.49, R) >droWil1.scaffold_180708 11651640 91 + 12563649 GUCUUCAACGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCUGA--GUUAAGAU-- (((((.((((((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))...--))))))))-- ( -27.90, z-score = -1.65, R) >droPer1.super_1 10242609 91 - 10282868 GUUCAAAGCGGUUCUAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUAGAACCUGA--AGUGAAAU-- .((((...((((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))).).--..))))..-- ( -26.20, z-score = -1.83, R) >dp4.chr4_group3 8790860 91 - 11692001 GUUCAAAGCGGUUCUAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUAGAACCUGA--AGUGAAAU-- .((((...((((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))).).--..))))..-- ( -26.20, z-score = -1.83, R) >droEre2.scaffold_4929 2056848 91 - 26641161 GCUCAUAGAGGUUCUAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUAGAACCUAU--ACGGACUU-- (..(.((.((((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))).)--).)..)..-- ( -25.50, z-score = -1.06, R) >droYak2.chr2L 1995823 91 - 22324452 GCUCAUAGAGGUUCUAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUAGAACCUAU--ACUAAGUU-- (((.....((((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))))..--....))).-- ( -25.10, z-score = -1.35, R) >droSec1.super_14 1957687 91 - 2068291 ACCCAUAGAGGUUCUAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUAGAACCUAG--ACUAAAUU-- ........((((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))))..--........-- ( -24.90, z-score = -1.32, R) >droSim1.chr2L 1980529 86 - 22036055 -----UAGAGGUUCUAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUAGAACCUAG--ACUACAUU-- -----...((((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))))..--........-- ( -24.90, z-score = -1.46, R) >consensus G_UCAUAGAGGUUCUAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUAGAACCUAA__ACUAAAUU__ .........(((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))............... (-24.79 = -24.33 + -0.46)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:54 2011