
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,768,066 – 21,768,131 |
| Length | 65 |
| Max. P | 0.630947 |

| Location | 21,768,066 – 21,768,131 |
|---|---|
| Length | 65 |
| Sequences | 9 |
| Columns | 73 |
| Reading direction | forward |
| Mean pairwise identity | 81.62 |
| Shannon entropy | 0.34976 |
| G+C content | 0.55462 |
| Mean single sequence MFE | -25.41 |
| Consensus MFE | -16.66 |
| Energy contribution | -16.33 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.630947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 21768066 65 + 23011544 -----GGUGCGGCAGGUGCGAAAUGUAUCCUGCUCCUCAUUGUCGGCUGCACGAAGAUG--CAGGAUGUUCG- -----.(.(((((((((((.....))).)))))((((((((.(((......))).))))--.))))))).).- ( -21.20, z-score = -0.54, R) >droSim1.chr2L 21311445 65 + 22036055 -----GGUGCGGCAGGUGCGAAAUGUAUCCUGCUGCUCAUUGUCGGCUGCACGAAGAUG--CAGGAUGUUUG- -----.(.(((((((((((.....))).)))))))).)........(((((......))--)))........- ( -23.10, z-score = -1.22, R) >droSec1.super_28 328106 64 + 873875 -----GGUGCGGCAGGUGCGAAAUGUAUCCUGCUGCUCAUUGUCGGCUGCACGAAGAUG--CAGGAUGUUU-- -----.(.(((((((((((.....))).)))))))).)........(((((......))--))).......-- ( -23.10, z-score = -1.29, R) >droYak2.chr2R 20062638 65 - 21139217 -----GGUGCGGCAGGUGCGAAAUGUUUCCUGCUGCUCAUUGUCGGCUGCACGGAGAUG--CAGAAUUUUCA- -----.(.((((((((.((.....))..)))))))).)........(((((......))--)))........- ( -21.50, z-score = -0.58, R) >droEre2.scaffold_4845 19825624 65 - 22589142 -----GGUGCGGCAGGUGCGAAAUGUAUCCUGCCGCUCAUUGUCGGCUGCACGAAGAUG--CAGACUGUUCG- -----.(.(((((((((((.....))).)))))))).).....((((((((......))--))).)))....- ( -27.60, z-score = -2.43, R) >droAna3.scaffold_12943 2005966 67 - 5039921 -----GGUGCGGCAGGUGCAAAAUGUAUCCUGCUGCUCAUUGUCAGCUGCACAAGGAUGUGCAGGAUGCAGG- -----.(.(((((((((((.....))).)))))))).).(((.((.(((((((....)))))))..))))).- ( -30.00, z-score = -2.85, R) >dp4.chr4_group1 1131140 70 + 5278887 GGUGCGGUGUGGCAGGUGCAAAAUGUAUCCUGCUGCUCAUUGUCAGCUGCAUAAGGAUG--CAGCA-GCAGGA ..(((.(.(..((((((((.....))).)))))..).).......(((((((....)))--)))).-)))... ( -29.80, z-score = -2.13, R) >droPer1.super_5 2736218 70 - 6813705 GGUGCGGUGUGGCAGGUGCAAAAUGUAUCCUGCUGCUCAUUGUCAGCUGCAUAAGGAUG--CAGCA-GCAGGA ..(((.(.(..((((((((.....))).)))))..).).......(((((((....)))--)))).-)))... ( -29.80, z-score = -2.13, R) >droVir3.scaffold_12963 14637993 66 - 20206255 -----GCUGCGGCAGGUGC-AAAUGUAUCCUGCUGCUCAUUGUCAGCUGCAUGCGAAGGUGCCACUUUCUCG- -----...(((((((((((-....))).)))))))).........((.....))(((((.....)))))...- ( -22.60, z-score = -0.78, R) >consensus _____GGUGCGGCAGGUGCGAAAUGUAUCCUGCUGCUCAUUGUCGGCUGCACGAAGAUG__CAGGAUGUUCG_ ......(((((((((((((.....))).))))))(((.......))).))))..................... (-16.66 = -16.33 + -0.33)
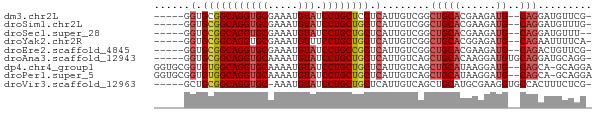
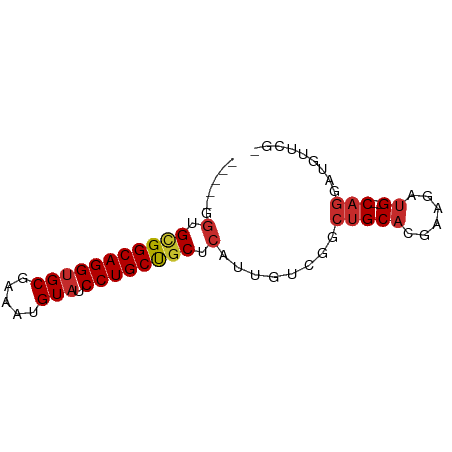

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:56:59 2011