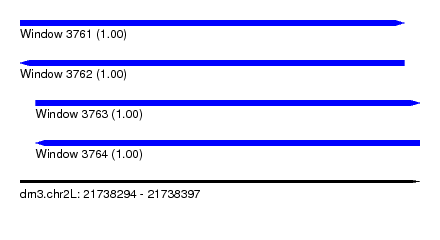
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,738,294 – 21,738,397 |
| Length | 103 |
| Max. P | 0.999837 |

| Location | 21,738,294 – 21,738,393 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 60.87 |
| Shannon entropy | 0.54627 |
| G+C content | 0.37540 |
| Mean single sequence MFE | -23.80 |
| Consensus MFE | -13.42 |
| Energy contribution | -14.10 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.41 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.29 |
| SVM RNA-class probability | 0.999739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
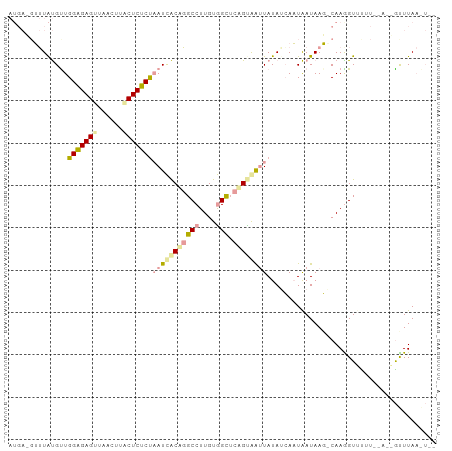
>dm3.chr2L 21738294 99 + 23011544 CGACUUAAGCCGU--AAAAACCUUGUCUUAUUAUUGAUAUAAUUGCCGAGCCACAAGGCCGGUGAUUAGAGAGGAAGUAAACUCUCCAGCAUAAACGUCAU .((((((.((...--........((((........))))((((..(((.(((....))))))..))))(((((........)))))..)).)))..))).. ( -27.70, z-score = -3.40, R) >droSim1.chrU 4996046 83 - 15797150 -----UAAAU------AAAACCUUG-CUUAUCAUUGAUAUAAUUACUGAGCUACAAGGCGUGCAAUUGGGGAGUAAAGUAACUCCCCAACGCACA------ -----.....------....(((((-....(((.((((...)))).)))....))))).((((..(((((((((......))))))))).)))).------ ( -28.70, z-score = -5.26, R) >droPer1.super_1039 3480 98 - 6997 --AUUUAAACACUUUAAGCACUUUGAAUUAUUAAUUCUAUA-CCACUGUACCACAAGGCCAGUGAUAAGAGAGACAGUUAUCUCUCCAACAUAAUCUUCAU --.....................((((..((((........-.(((((..((....)).)))))....((((((......)))))).....)))).)))). ( -15.00, z-score = -1.56, R) >consensus __A_UUAAAC__U__AAAAACCUUG_CUUAUUAUUGAUAUAAUUACUGAGCCACAAGGCCAGUGAUUAGAGAGAAAGUUAACUCUCCAACAUAAAC_UCAU .......................................(((((((((.(((....))))))))))))(((((........)))))............... (-13.42 = -14.10 + 0.68)

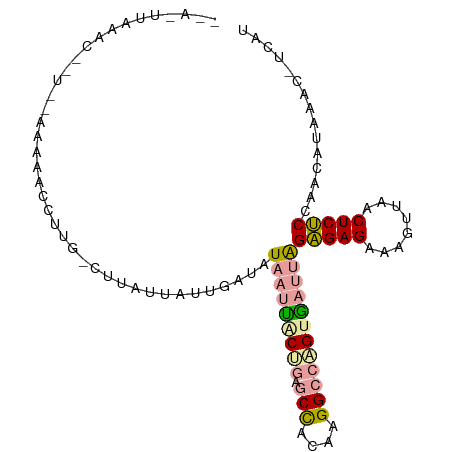
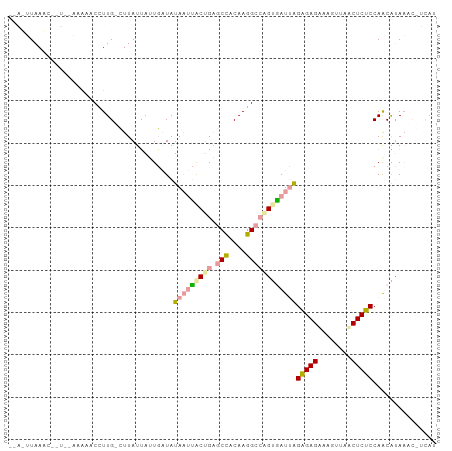
| Location | 21,738,294 – 21,738,393 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 60.87 |
| Shannon entropy | 0.54627 |
| G+C content | 0.37540 |
| Mean single sequence MFE | -27.64 |
| Consensus MFE | -15.64 |
| Energy contribution | -16.43 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.44 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.22 |
| SVM RNA-class probability | 0.999705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
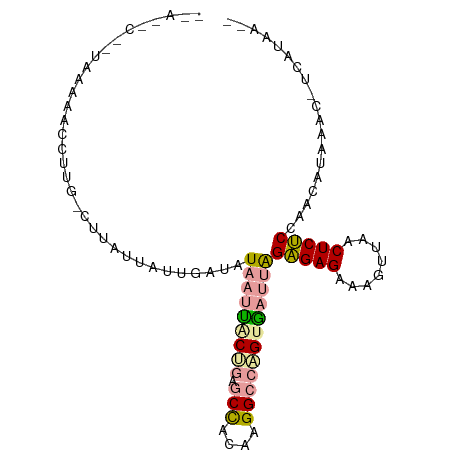
>dm3.chr2L 21738294 99 - 23011544 AUGACGUUUAUGCUGGAGAGUUUACUUCCUCUCUAAUCACCGGCCUUGUGGCUCGGCAAUUAUAUCAAUAAUAAGACAAGGUUUUU--ACGGCUUAAGUCG ..(((......(((((((((........)))))((((..((((((....))).)))..))))........................--.))))....))). ( -24.70, z-score = -1.57, R) >droSim1.chrU 4996046 83 + 15797150 ------UGUGCGUUGGGGAGUUACUUUACUCCCCAAUUGCACGCCUUGUAGCUCAGUAAUUAUAUCAAUGAUAAG-CAAGGUUUU------AUUUA----- ------.((((((((((((((......))))))))).)))))(((((((...(((.............)))...)-))))))...------.....----- ( -32.12, z-score = -5.49, R) >droPer1.super_1039 3480 98 + 6997 AUGAAGAUUAUGUUGGAGAGAUAACUGUCUCUCUUAUCACUGGCCUUGUGGUACAGUGG-UAUAGAAUUAAUAAUUCAAAGUGCUUAAAGUGUUUAAAU-- .((((......(((((((((((....))))))))(((((((((((....))).))))))-))...)))......))))(((..(.....)..)))....-- ( -26.10, z-score = -2.86, R) >consensus AUGA_GUUUAUGUUGGAGAGUUAACUUACUCUCUAAUCACAGGCCUUGUGGCUCAGUAAUUAUAUCAAUAAUAAG_CAAGGUUUUU__A__GUUUAA_U__ ..............(((((((......)))))))(((((((((((....))).))))))))........................................ (-15.64 = -16.43 + 0.79)

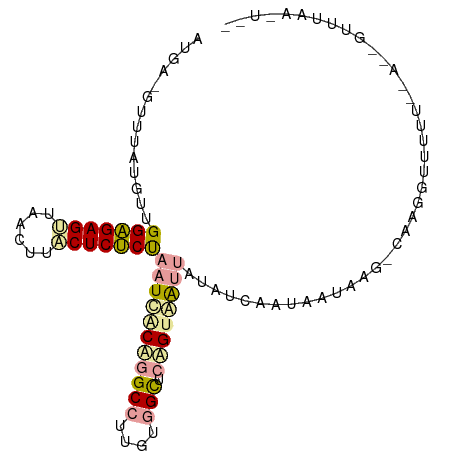
| Location | 21,738,298 – 21,738,397 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 58.45 |
| Shannon entropy | 0.59351 |
| G+C content | 0.37461 |
| Mean single sequence MFE | -23.23 |
| Consensus MFE | -13.42 |
| Energy contribution | -14.10 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.41 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.53 |
| SVM RNA-class probability | 0.999837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
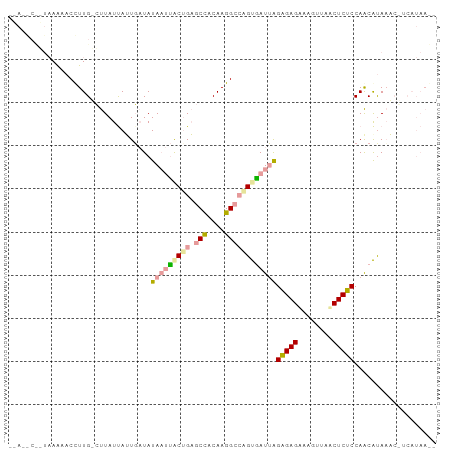
>dm3.chr2L 21738298 99 + 23011544 UUAAGCCGUAAAAACCUUGUCUUAUUAUUGAUAUAAUUGCCGAGCCACAAGGCCGGUGAUUAGAGAGGAAGUAAACUCUCCAGCAUAAACGUCAUAAGA ...................(((((....((((.((((..(((.(((....))))))..))))(((((........)))))..........))))))))) ( -26.00, z-score = -3.04, R) >droSim1.chrU 4996049 80 - 15797150 --------AUAAAACCUUG-CUUAUCAUUGAUAUAAUUACUGAGCUACAAGGCGUGCAAUUGGGGAGUAAAGUAACUCCCCAACGCACA---------- --------......(((((-....(((.((((...)))).)))....))))).((((..(((((((((......))))))))).)))).---------- ( -28.70, z-score = -5.23, R) >droPer1.super_1039 3484 97 - 6997 AAACACUUUAAGCACUUUGAAUUAUUAAUUCUAUA-CCACUGUACCACAAGGCCAGUGAUAAGAGAGACAGUUAUCUCUCCAACAUAAUCUUCAUAAA- .................((((..((((........-.(((((..((....)).)))))....((((((......)))))).....)))).))))....- ( -15.00, z-score = -1.61, R) >consensus __A__C__UAAAAACCUUG_CUUAUUAUUGAUAUAAUUACUGAGCCACAAGGCCAGUGAUUAGAGAGAAAGUUAACUCUCCAACAUAAAC_UCAUAA__ .................................(((((((((.(((....))))))))))))(((((........)))))................... (-13.42 = -14.10 + 0.68)

| Location | 21,738,298 – 21,738,397 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 58.45 |
| Shannon entropy | 0.59351 |
| G+C content | 0.37461 |
| Mean single sequence MFE | -26.61 |
| Consensus MFE | -15.64 |
| Energy contribution | -16.43 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.44 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.33 |
| SVM RNA-class probability | 0.999762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
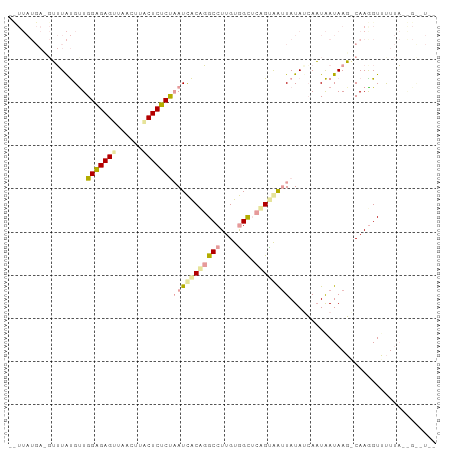
>dm3.chr2L 21738298 99 - 23011544 UCUUAUGACGUUUAUGCUGGAGAGUUUACUUCCUCUCUAAUCACCGGCCUUGUGGCUCGGCAAUUAUAUCAAUAAUAAGACAAGGUUUUUACGGCUUAA ((((((......((((.(((((((........)))))))....((((((....))).)))....))))......))))))..(((((.....))))).. ( -22.30, z-score = -1.09, R) >droSim1.chrU 4996049 80 + 15797150 ----------UGUGCGUUGGGGAGUUACUUUACUCCCCAAUUGCACGCCUUGUAGCUCAGUAAUUAUAUCAAUGAUAAG-CAAGGUUUUAU-------- ----------.((((((((((((((......))))))))).)))))(((((((...(((.............)))...)-)))))).....-------- ( -32.12, z-score = -5.59, R) >droPer1.super_1039 3484 97 + 6997 -UUUAUGAAGAUUAUGUUGGAGAGAUAACUGUCUCUCUUAUCACUGGCCUUGUGGUACAGUGG-UAUAGAAUUAAUAAUUCAAAGUGCUUAAAGUGUUU -....((((......(((((((((((....))))))))(((((((((((....))).))))))-))...)))......))))(((..(.....)..))) ( -25.40, z-score = -2.51, R) >consensus __UUAUGA_GUUUAUGUUGGAGAGUUAACUUACUCUCUAAUCACAGGCCUUGUGGCUCAGUAAUUAUAUCAAUAAUAAG_CAAGGUUUUUA__G__U__ ..................(((((((......)))))))(((((((((((....))).)))))))).................................. (-15.64 = -16.43 + 0.79)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:56:57 2011