| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,702,653 – 21,702,791 |
| Length | 138 |
| Max. P | 0.925142 |

| Location | 21,702,653 – 21,702,791 |
|---|---|
| Length | 138 |
| Sequences | 4 |
| Columns | 142 |
| Reading direction | forward |
| Mean pairwise identity | 78.61 |
| Shannon entropy | 0.35130 |
| G+C content | 0.35627 |
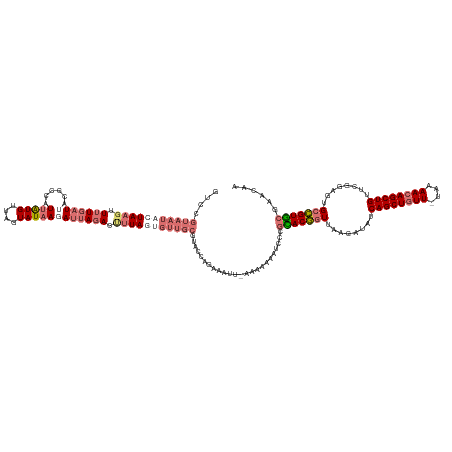
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -17.66 |
| Energy contribution | -17.72 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.708546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
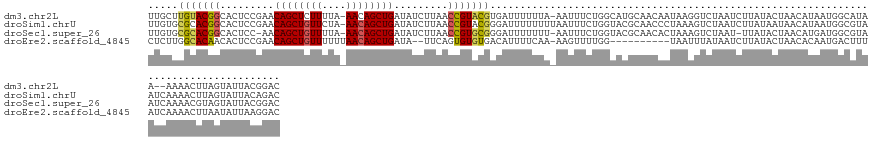
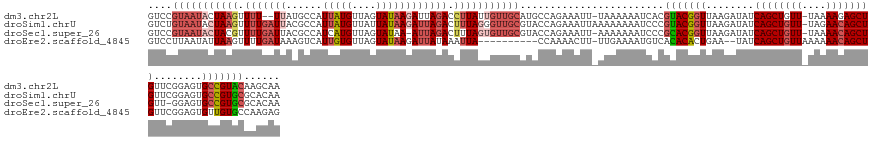
>dm3.chr2L 21702653 138 + 23011544 UUGCUUGUACGGCACUCCGAACAGCUCUUUUA-AACAGCUGAUAUCUUAACCGUACGUGAUUUUUUA-AAUUUCUGGCAUGCAACAAUAAGGUCUAAUCUUAUACUAACAUAAUGGCAUAA--AAAACUUAGUAUUACGGAC ......((((((.........(((((......-...))))).........))))))...........-....((((..((((....(((((((...))))))).(((......))).....--........))))..)))). ( -21.77, z-score = 0.03, R) >droSim1.chrU 4924553 141 - 15797150 UUGUGCGCACGGCACUCCGAACAGCUGUUCUA-AACAGCUGAUAUCUUAACCGUACGGGAUUUUUUUUAAUUUCUGGUACGCAACCCUAAAGUCUAAUCUUAUAAUAACAUAAUGGCGUAAUCAAAACUUAGUAUUACAGAC ...(((((.(((....)))..(((((((....-.)))))))..........(((((.(((..((....))..))).)))))..................................)))))...................... ( -28.80, z-score = -1.09, R) >droSec1.super_26 339118 138 + 826190 UUGUGCGCACGGCACUCC-AACAGCUGUUUUA-AACAGCUGAUAUCUUAACCGUGCGGGAUUUUUUU-AAUUUCUGGUACGCAACACUAAAGUCUAAU-UUAUACUAACAUGAUGGCGUAAUCAAAACGUAGUAUUACGGAC ..((((((((((......-..(((((((....-.))))))).........))))))(..(((.....-)))..)..))))...........((((...-..((((((...((((......)))).....))))))...)))) ( -36.13, z-score = -2.52, R) >droEre2.scaffold_4845 20085830 129 - 22589142 CUCUUGGCACAACACUCCGAACAGCUGUUUUUUAACAGCUGAUA--UUCAGUGUGUGACAUUUUCAA-AAGUUUUGG----------UAAUUUAUAAUCUUAUACUAACACAAUGACUUUAUCAAAACUUAAUAUUAAGGAC .((((((((((.((((..(((((((((((....))))))))...--)))))))))))..........-(((((((((----------(((.((((.................))))..))))))))))))....)))))).. ( -30.63, z-score = -3.82, R) >consensus UUGUGCGCACGGCACUCCGAACAGCUGUUUUA_AACAGCUGAUAUCUUAACCGUACGGGAUUUUUUA_AAUUUCUGGUACGCAACACUAAAGUCUAAUCUUAUACUAACAUAAUGGCGUAAUCAAAACUUAGUAUUACGGAC .....(((((((.........((((((((....)))))))).........)))))))..................................................................................... (-17.66 = -17.72 + 0.06)
| Location | 21,702,653 – 21,702,791 |
|---|---|
| Length | 138 |
| Sequences | 4 |
| Columns | 142 |
| Reading direction | reverse |
| Mean pairwise identity | 78.61 |
| Shannon entropy | 0.35130 |
| G+C content | 0.35627 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -22.75 |
| Energy contribution | -27.19 |
| Covariance contribution | 4.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.925142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
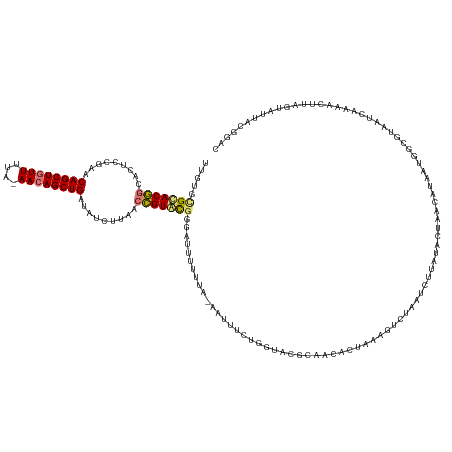
>dm3.chr2L 21702653 138 - 23011544 GUCCGUAAUACUAAGUUUU--UUAUGCCAUUAUGUUAGUAUAAGAUUAGACCUUAUUGUUGCAUGCCAGAAAUU-UAAAAAAUCACGUACGGUUAAGAUAUCAGCUGUU-UAAAAGAGCUGUUCGGAGUGCCGUACAAGCAA ....((((((.((((..((--((((((.(......).))))))))......)))).)))))).(((..((....-.......))..(((((((...((...(((((.(.-....).))))).)).....)))))))..))). ( -29.80, z-score = -1.22, R) >droSim1.chrU 4924553 141 + 15797150 GUCUGUAAUACUAAGUUUUGAUUACGCCAUUAUGUUAUUAUAAGAUUAGACUUUAGGGUUGCGUACCAGAAAUUAAAAAAAAUCCCGUACGGUUAAGAUAUCAGCUGUU-UAGAACAGCUGUUCGGAGUGCCGUGCGCACAA ((.(((((..(((((.(((((((......(((((....)))))))))))).)))))..))))).))...................((((((((...((...(((((((.-....))))))).)).....))))))))..... ( -35.70, z-score = -1.73, R) >droSec1.super_26 339118 138 - 826190 GUCCGUAAUACUACGUUUUGAUUACGCCAUCAUGUUAGUAUAA-AUUAGACUUUAGUGUUGCGUACCAGAAAUU-AAAAAAAUCCCGCACGGUUAAGAUAUCAGCUGUU-UAAAACAGCUGUU-GGAGUGCCGUGCGCACAA ((.((((((((((.(.(((((((((............)))...-)))))).).)))))))))).))........-..........((((((((.....((.(((((((.-....))))))).)-)....))))))))..... ( -38.60, z-score = -2.92, R) >droEre2.scaffold_4845 20085830 129 + 22589142 GUCCUUAAUAUUAAGUUUUGAUAAAGUCAUUGUGUUAGUAUAAGAUUAUAAAUUA----------CCAAAACUU-UUGAAAAUGUCACACACUGAA--UAUCAGCUGUUAAAAAACAGCUGUUCGGAGUGUUGUGCCAAGAG ............((((((((.(((((((.(((((....))))))))).....)))----------.))))))))-.......((.((((((((...--...((((((((....)))))))).....)))).)))).)).... ( -27.90, z-score = -2.05, R) >consensus GUCCGUAAUACUAAGUUUUGAUUACGCCAUUAUGUUAGUAUAAGAUUAGACUUUAGUGUUGCGUACCAGAAAUU_AAAAAAAUCCCGCACGGUUAAGAUAUCAGCUGUU_UAAAACAGCUGUUCGGAGUGCCGUGCGAACAA ((.((((((((((((.(((((((......(((((....)))))))))))).)))))))))))).))....................(((((((........((((((((....))))))))........)))))))...... (-22.75 = -27.19 + 4.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:56:53 2011