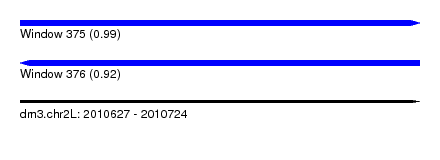
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,010,627 – 2,010,724 |
| Length | 97 |
| Max. P | 0.992814 |

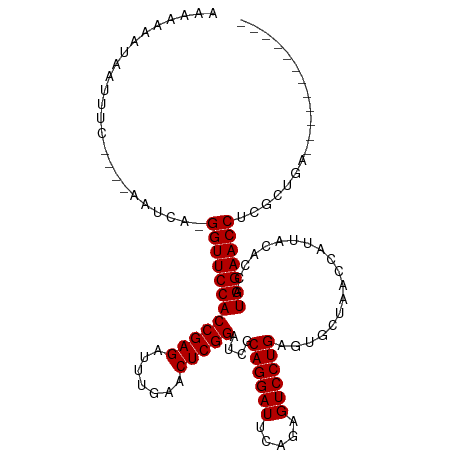
| Location | 2,010,627 – 2,010,724 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 83.72 |
| Shannon entropy | 0.33719 |
| G+C content | 0.45159 |
| Mean single sequence MFE | -28.84 |
| Consensus MFE | -25.79 |
| Energy contribution | -25.79 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.992814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
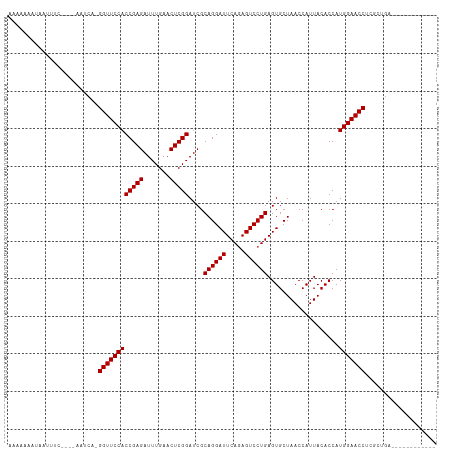
>dm3.chr2L 2010627 97 + 23011544 AAAAAACUAAUUUC----AUUCA-GGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCUCGCUGA------------ ............((----(.(.(-((((((((((((.......))))).....((.((((((....)))))).)).............)))))))).).)))------------ ( -27.90, z-score = -2.61, R) >droSim1.chr2L 1980295 97 + 22036055 AAAAAAAUAAUUUC----AUUCA-GGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCUAGCUGA------------ ............((----(.(.(-((((((((((((.......))))).....((.((((((....)))))).)).............)))))))).).)))------------ ( -27.90, z-score = -2.88, R) >droSec1.super_14 1957456 97 + 2068291 AAAAAAGUAAUUUC----AAUCA-GGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCUCGCUGA------------ ............((----(.(.(-((((((((((((.......))))).....((.((((((....)))))).)).............)))))))).).)))------------ ( -27.90, z-score = -2.35, R) >droYak2.chr2L 1995585 97 + 22324452 AAAAAAGUAAUCUC----AAUCA-GGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCUCGCUGA------------ ............((----(.(.(-((((((((((((.......))))).....((.((((((....)))))).)).............)))))))).).)))------------ ( -27.80, z-score = -2.16, R) >droEre2.scaffold_4929 2056613 93 + 26641161 -AAAAAAUGAACU-------UCA-GGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCUCACUGA------------ -............-------..(-((((((((((((.......))))).....((.((((((....)))))).)).............))))))))......------------ ( -27.00, z-score = -2.39, R) >dp4.chr4_group3 8790607 102 + 11692001 AAAAAAAAGAUUUUCAGAAAUCAAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCUCUCUGA------------ .............(((((.....(((((((((((((.......))))).....((.((((((....)))))).)).............)))))))).)))))------------ ( -30.40, z-score = -2.76, R) >droPer1.super_1 10242358 102 + 10282868 AAAAAAAAGAUUUUCAGAAAUCAAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCUCUCUGA------------ .............(((((.....(((((((((((((.......))))).....((.((((((....)))))).)).............)))))))).)))))------------ ( -30.40, z-score = -2.76, R) >droWil1.scaffold_180708 11651337 110 - 12563649 AAAAAGCUAAUUUUC---GAUCA-GGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAAAGUCCUGAGUGCUAACCAUUACACCAUGGAACCGGGCUGGAAAUGUGGCCGA .....((((((((((---(..(.-((((((((((((.......)))))....((((((.....))))))...................))))))).)..))))))).))))... ( -36.60, z-score = -2.56, R) >droVir3.scaffold_12963 7606855 93 - 20206255 -----AAGAAUU--CG-AACUCA-GGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCCAUACGA------------ -----.......--..-......-((((((((((((.......))))).....((.((((((....)))))).)).............))))))).......------------ ( -26.00, z-score = -2.11, R) >droMoj3.scaffold_6500 29330731 101 + 32352404 AAAAAUUAGACUGAUAUAGUCCA-GGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCGAUGCGA------------ ........(((((...)))))..-((((((((((((.......))))).....((.((((((....)))))).)).............))))))).......------------ ( -29.40, z-score = -2.56, R) >droGri2.scaffold_15252 165374 96 + 17193109 ----GAAAGAUCGCUA-AGCUCA-GGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCCAUAUGG------------ ----............-......-((((((((((((.......))))).....((.((((((....)))))).)).............))))))).......------------ ( -26.00, z-score = -1.32, R) >anoGam1.chr3R 22019152 102 - 53272125 AAAAAAGUGACUCG----GGACA-GGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAAAGUCCUGAGUGCUAACCAUUACACCAUGGAACCGUAUGCAAGUUG------- ........((((.(----.(...-((((((((((((.......)))))....((((((.....))))))...................)))))))...).).)))).------- ( -28.80, z-score = -1.56, R) >consensus AAAAAAAUAAUUUC____AAUCA_GGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCUCGCUGA____________ ........................((((((((((((.......)))))....((((((.....))))))...................)))))))................... (-25.79 = -25.79 + 0.00)

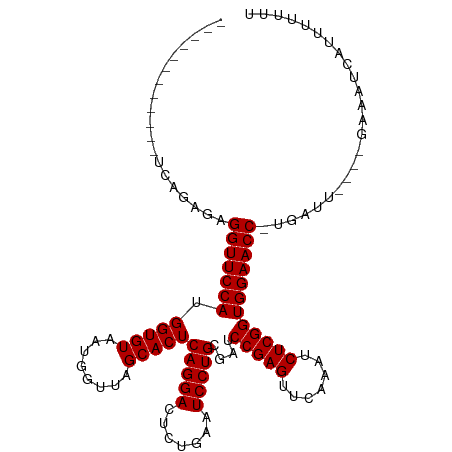
| Location | 2,010,627 – 2,010,724 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 83.72 |
| Shannon entropy | 0.33719 |
| G+C content | 0.45159 |
| Mean single sequence MFE | -29.44 |
| Consensus MFE | -24.59 |
| Energy contribution | -24.59 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2010627 97 - 23011544 ------------UCAGCGAGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACC-UGAAU----GAAAUUAGUUUUUU ------------(((...((((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))-)...)----))............ ( -28.80, z-score = -1.62, R) >droSim1.chr2L 1980295 97 - 22036055 ------------UCAGCUAGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACC-UGAAU----GAAAUUAUUUUUUU ------------(((..(((((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))-))..)----))............ ( -30.70, z-score = -2.72, R) >droSec1.super_14 1957456 97 - 2068291 ------------UCAGCGAGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACC-UGAUU----GAAAUUACUUUUUU ------------((((..((((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))-)..))----))............ ( -29.80, z-score = -1.98, R) >droYak2.chr2L 1995585 97 - 22324452 ------------UCAGCGAGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACC-UGAUU----GAGAUUACUUUUUU ------------((((..((((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))-)..))----))............ ( -29.80, z-score = -1.55, R) >droEre2.scaffold_4929 2056613 93 - 26641161 ------------UCAGUGAGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACC-UGA-------AGUUCAUUUUUU- ------------....(.((((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))-).)-------............- ( -27.20, z-score = -1.22, R) >dp4.chr4_group3 8790607 102 - 11692001 ------------UCAGAGAGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCUUGAUUUCUGAAAAUCUUUUUUUU ------------((((((((((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))))....)))))............. ( -32.30, z-score = -2.22, R) >droPer1.super_1 10242358 102 - 10282868 ------------UCAGAGAGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCUUGAUUUCUGAAAAUCUUUUUUUU ------------((((((((((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))))....)))))............. ( -32.30, z-score = -2.22, R) >droWil1.scaffold_180708 11651337 110 + 12563649 UCGGCCACAUUUCCAGCCCGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUUUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACC-UGAUC---GAAAAUUAGCUUUUU (((((..........))).(((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))-.....---))............. ( -28.10, z-score = -0.14, R) >droVir3.scaffold_12963 7606855 93 + 20206255 ------------UCGUAUGGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACC-UGAGUU-CG--AAUUCUU----- ------------(((..(((((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))-))....-))--)......----- ( -28.10, z-score = -1.29, R) >droMoj3.scaffold_6500 29330731 101 - 32352404 ------------UCGCAUCGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACC-UGGACUAUAUCAGUCUAAUUUUU ------------.......(((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))-((((((.....))))))...... ( -30.60, z-score = -2.01, R) >droGri2.scaffold_15252 165374 96 - 17193109 ------------CCAUAUGGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACC-UGAGCU-UAGCGAUCUUUC---- ------------......((((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))-).....-............---- ( -26.70, z-score = -0.44, R) >anoGam1.chr3R 22019152 102 + 53272125 -------CAACUUGCAUACGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUUUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACC-UGUCC----CGAGUCACUUUUUU -------..(((((...(((((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))-.))..----)))))......... ( -28.90, z-score = -1.32, R) >consensus ____________UCAGAGAGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACC_UGAUU____GAAAUCAUUUUUUU ...................(((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))........................ (-24.59 = -24.59 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:52 2011