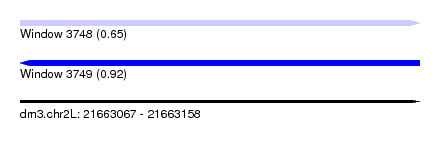
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,663,067 – 21,663,158 |
| Length | 91 |
| Max. P | 0.920983 |

| Location | 21,663,067 – 21,663,158 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 73.97 |
| Shannon entropy | 0.48717 |
| G+C content | 0.28265 |
| Mean single sequence MFE | -17.82 |
| Consensus MFE | -7.28 |
| Energy contribution | -7.39 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.648889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
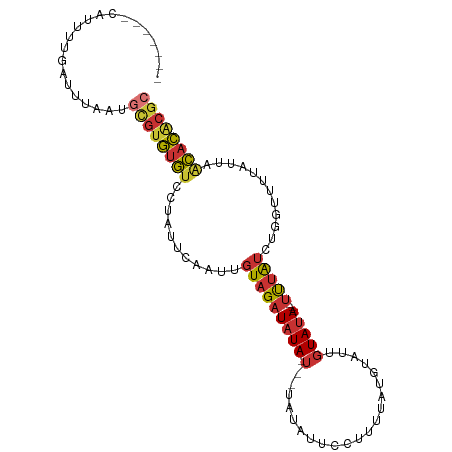
>dm3.chr2L 21663067 91 + 23011544 -------CAUUUUGAUUUAAUGCGUGUGUCCUAUUCAAUUGUAGAUAUA-A--UAUAUUUCUUU-AUGUAUUGUAUA---UUUAUCUGGUUUUAUUAACACACGC -------..............((((((((.((((......))))(((((-(--(((((......-))))))))))).---.................)))))))) ( -21.10, z-score = -3.53, R) >droSim1.chr2L 21215570 92 + 22036055 -------CAUUUUGAUUUAAUGCGUGUGUCCUAUUCAAUUGUAGAUAUA-U--UAUAUUGCUUUUAUGUAUUGUAUA---UUUAUCUGGUUUUAUUAACACACGC -------..............((((((((...((..(((((((((((((-(--.((((.........)))).)))))---)))))..))))..))..)))))))) ( -19.10, z-score = -2.32, R) >droSec1.super_28 206376 92 + 873875 -------CAUUUUGAUUUAAUGCGUGUGUCCUAUUCAAUUGUAGAUAUA-U--UAUAUUGCUUUUAUGUAUUGUAUA---UUUAUCUGGUUUUAUUAACACACGC -------..............((((((((...((..(((((((((((((-(--.((((.........)))).)))))---)))))..))))..))..)))))))) ( -19.10, z-score = -2.32, R) >droYak2.chr2R 19958787 92 - 21139217 -------CAUUUUGAUUUAAUGCGUGUGUCCUAUUCAAUUGUAGAUAUA-UAUUAUAUUCCUUUUAUGUAUUGUAUA---UUUAUCUGGUUUUAUUCACACGC-- -------..............(((((((......(((...(((((((((-((.(((((.......))))).))))))---))))).))).......)))))))-- ( -21.62, z-score = -4.12, R) >droAna3.scaffold_12916 9261814 87 + 16180835 -------CGUUUUGAUUUAAUGCGUGCGUCCAUUAUAUUUGUAGAUAUA-U-------UCCUUUUGUGUAAUGUAUA---UUUAUCUGGUUUUAUGCGCACACGC -------..............(((((.((.(((...(((.(((((((((-(-------..............)))))---)))))..)))...))).)).))))) ( -16.94, z-score = -0.88, R) >dp4.chr4_group1 4057332 88 + 5278887 -------CGUUUUGAUUUAAUGCGUGUGUUUUAUAUUAUUGUAGAUAUA-U------CCCAUUUUGUGUAUUGUAUA---UUUAUCUGGUUUUAUGCGCACACGC -------..............((((((((..((((..((((((((((((-(------..(((...)))....)))))---)))))..)))..)))).)))))))) ( -18.30, z-score = -1.63, R) >droPer1.super_5 5601294 88 - 6813705 -------CGUUUUGAUUUAAUGCGUGUGUUUUAUAUUAUUGUAGAUAUA-U------CCCAUUUUGUGUAUUGUAUA---UUUAUCUGGUUUUAUGCGCACACGC -------..............((((((((..((((..((((((((((((-(------..(((...)))....)))))---)))))..)))..)))).)))))))) ( -18.30, z-score = -1.63, R) >triCas2.ChLG7 3081652 104 + 17478683 CAAUCUGAGUUCUGGCGCUCUCUUGAUAUUCCUUUGAAUUUUUGUUUUAAUUCUGCUUUUACUUUAUUAAUUGUAUAGAAUCUGUUUAUUUGUAUUACUAUGUG- ......((((......))))...((((((.....(((((....((((((....(((..(((......)))..)))))))))..)))))...)))))).......- ( -8.10, z-score = 1.11, R) >consensus _______CAUUUUGAUUUAAUGCGUGUGUCCUAUUCAAUUGUAGAUAUA_U__UAUAUUCCUUUUAUGUAUUGUAUA___UUUAUCUGGUUUUAUUAACACACGC .....................((((((((............((((((...................................)))))).........)))))))) ( -7.28 = -7.39 + 0.11)

| Location | 21,663,067 – 21,663,158 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 73.97 |
| Shannon entropy | 0.48717 |
| G+C content | 0.28265 |
| Mean single sequence MFE | -13.34 |
| Consensus MFE | -5.31 |
| Energy contribution | -5.64 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
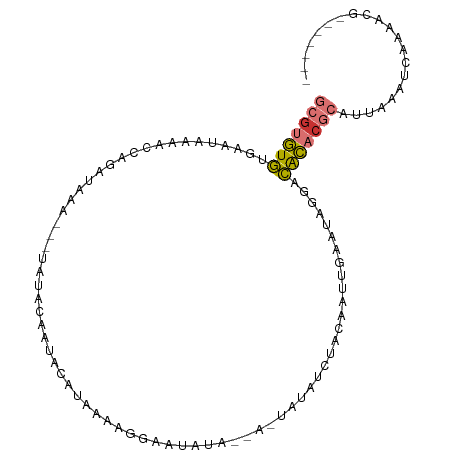
>dm3.chr2L 21663067 91 - 23011544 GCGUGUGUUAAUAAAACCAGAUAAA---UAUACAAUACAU-AAAGAAAUAUA--U-UAUAUCUACAAUUGAAUAGGACACACGCAUUAAAUCAAAAUG------- (((((((((.........(((((((---((((........-.......))))--)-).)))))............)))))))))..............------- ( -17.36, z-score = -4.06, R) >droSim1.chr2L 21215570 92 - 22036055 GCGUGUGUUAAUAAAACCAGAUAAA---UAUACAAUACAUAAAAGCAAUAUA--A-UAUAUCUACAAUUGAAUAGGACACACGCAUUAAAUCAAAAUG------- (((((((((.........(((((..---((((................))))--.-..)))))............)))))))))..............------- ( -14.49, z-score = -2.69, R) >droSec1.super_28 206376 92 - 873875 GCGUGUGUUAAUAAAACCAGAUAAA---UAUACAAUACAUAAAAGCAAUAUA--A-UAUAUCUACAAUUGAAUAGGACACACGCAUUAAAUCAAAAUG------- (((((((((.........(((((..---((((................))))--.-..)))))............)))))))))..............------- ( -14.49, z-score = -2.69, R) >droYak2.chr2R 19958787 92 + 21139217 --GCGUGUGAAUAAAACCAGAUAAA---UAUACAAUACAUAAAAGGAAUAUAAUA-UAUAUCUACAAUUGAAUAGGACACACGCAUUAAAUCAAAAUG------- --(((((((.........(((((.(---(((......(......).......)))-).))))).(....).......)))))))..............------- ( -13.32, z-score = -2.60, R) >droAna3.scaffold_12916 9261814 87 - 16180835 GCGUGUGCGCAUAAAACCAGAUAAA---UAUACAUUACACAAAAGGA-------A-UAUAUCUACAAAUAUAAUGGACGCACGCAUUAAAUCAAAACG------- ((((((((.(((....((.......---................)).-------.-(((((......))))))))).)))))))..............------- ( -13.10, z-score = -1.74, R) >dp4.chr4_group1 4057332 88 - 5278887 GCGUGUGCGCAUAAAACCAGAUAAA---UAUACAAUACACAAAAUGGG------A-UAUAUCUACAAUAAUAUAAAACACACGCAUUAAAUCAAAACG------- (((((((.........(((......---................))).------.-(((((........)))))...)))))))..............------- ( -13.35, z-score = -2.71, R) >droPer1.super_5 5601294 88 + 6813705 GCGUGUGCGCAUAAAACCAGAUAAA---UAUACAAUACACAAAAUGGG------A-UAUAUCUACAAUAAUAUAAAACACACGCAUUAAAUCAAAACG------- (((((((.........(((......---................))).------.-(((((........)))))...)))))))..............------- ( -13.35, z-score = -2.71, R) >triCas2.ChLG7 3081652 104 - 17478683 -CACAUAGUAAUACAAAUAAACAGAUUCUAUACAAUUAAUAAAGUAAAAGCAGAAUUAAAACAAAAAUUCAAAGGAAUAUCAAGAGAGCGCCAGAACUCAGAUUG -.......................(((((..............((....)).(((((........)))))...))))).....(((..(....)..)))...... ( -7.30, z-score = -0.31, R) >consensus GCGUGUGUGAAUAAAACCAGAUAAA___UAUACAAUACAUAAAAGGAAUAUA__A_UAUAUCUACAAUUGAAUAGGACACACGCAUUAAAUCAAAACG_______ (((((((......................................................................)))))))..................... ( -5.31 = -5.64 + 0.33)

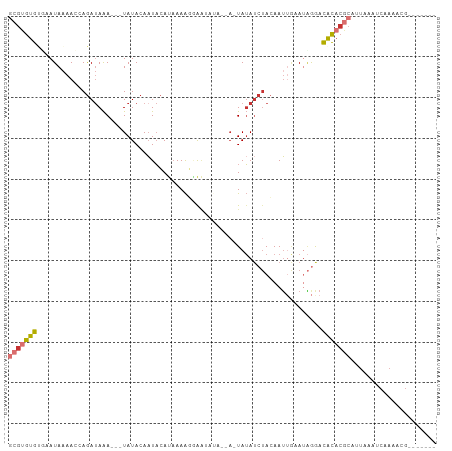
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:56:45 2011