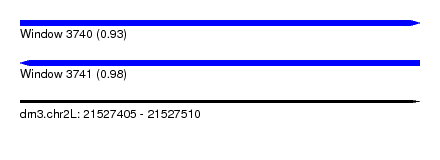
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,527,405 – 21,527,510 |
| Length | 105 |
| Max. P | 0.981098 |

| Location | 21,527,405 – 21,527,510 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 63.10 |
| Shannon entropy | 0.75309 |
| G+C content | 0.31909 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -6.03 |
| Energy contribution | -5.86 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.60 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934282 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 21527405 105 + 23011544 -UAUUUGGUACAUGUUCGCAAUUAAAAUUUU----AGAUUUAUGUUUUAUAGAUCUGAAAUUU--GUUUAAACAA-GUCCUUUUCAGGGCUACAACGUUCCGUUGC-AAGAGAA--- -................(((((..(((((((----(((((((((...))))))))))))))))--.........(-(((((....))))))..........)))))-.......--- ( -23.80, z-score = -2.38, R) >droSim1.chrU 3629637 105 - 15797150 -UAUUUGGUACAUGUUCGCAAUCAAAAAUUU----AGAUUUAUGAUUUAUAGAUCUGAAAUUU--GUUUAUACAA-GUCCUUUUCAGGGCUACAACGUUCUUUUGC-AAGAGAA--- -................((((.((((..(((----(((((((((...)))))))))))).)))--)........(-(((((....))))))...........))))-.......--- ( -19.20, z-score = -0.91, R) >droSec1.super_402 1611 105 + 12115 -UAUUUGGUACAUGUUCGCAAUCAAAACUUU----AGAUUUAUGAUUUAUAGAUCUGAAAUUU--GUUUAUACAA-GUCCUUUUCAGGGCUACAACGUUCUGUUGC-AAGAGAA--- -................((((.((.((((((----(((((((((...))))))))))))....--.........(-(((((....)))))).....))).))))))-.......--- ( -22.00, z-score = -1.42, R) >droYak2.chr2R_random 60132 98 - 67238 -UAGUUAGUACAUGUUCGCAAUCAAAAUUUU----AGAUUUCA-------ACAUCUGAAAUUU--GUUUAAACAA-GCCCUUUUCAGGGCUACAACGAUUCGUUGC-AAGAGAA--- -...........(((.((.((((.(((((((----((((....-------..)))))))))))--.........(-(((((....)))))).....))))))..))-)......--- ( -23.00, z-score = -2.89, R) >droEre2.scaffold_3221 10478 98 - 23538 -UAGUUGGUACAUGUUCGCAAUCAAAAUUUU----AGAUUUCU-------AGAUCUGAAAUUU--GUCUAAUCAA-GCCCUUUUCAGGGCUACAACAUUCACUUGC-AAGAGAA--- -..((.(((..(((((........(((((((----(((((...-------.))))))))))))--.........(-(((((....))))))..)))))..))).))-.......--- ( -24.40, z-score = -2.73, R) >droAna3.scaffold_13030 44304 99 - 47400 UAAAAUUGCACAUGUUUGCAAUCGAAAUUUU----AGAUUUCG-------AAAUCUAAAAUUA--AAUUAAACAA-GCCCUUUUCAGGGCUACAAG-AUAUGUUUUAAAGAGAA--- .........((((((((........((((((----(((((...-------.))))))))))).--.........(-(((((....))))))...))-))))))...........--- ( -22.40, z-score = -3.31, R) >dp4.Unknown_singleton_1517 15560 99 + 24252 -GUUGGUGUACAAGGUCGCAUUCGAAAUUUU----AGAUGUAU-------CCGUCUGAAAUUG--AUUUAAACAA-GCCCUUUUCAGGGCUACAAAACUAUAUUCAAAAGAGAU--- -(((..(((................((((((----(((((...-------.))))))))))).--.........(-(((((....))))))))).)))................--- ( -21.00, z-score = -1.36, R) >droPer1.super_0 3286671 99 + 11822988 -GUUGGUGUACAAGGUCGCAUUCGAAAUUUU----AGAUGUAU-------CCGUCUGAAAUUG--AUUUAAACAA-GCCCUUUUCAGGGCUACAAAACUAUAUUCAAAAGAGAU--- -(((..(((................((((((----(((((...-------.))))))))))).--.........(-(((((....))))))))).)))................--- ( -21.00, z-score = -1.36, R) >droWil1.scaffold_181036 31097 114 + 110856 --AAUUAAUACUUUAUUGCAAUCGUAAUAUUGAAAAAAUUUUAGAUAUCGAAUACGAAAAUCUAAAAUUUUACUUAAACACAGCCAGGUCGGC-GCGUUAAGUUUCAGCGUUAAGCG --...........((((((....)))))).((((((((((((((((.(((....)))..))))))))))))((((((.....(((.....)))-...))))))))))((.....)). ( -25.00, z-score = -2.61, R) >droVir3.scaffold_13047 1640872 100 + 19223366 --AAAUUGCUAACUGCU-UGUCAGCAUAUGC----AAAUUUCU-------AAUAAUGAAAUUUGUUUUCCUACAAAGCCCUUUUAAGGGCUACAAAUACGAUUUUCUAAGAGAA--- --....((((.((....-.)).))))...((----(((((((.-------......)))))))))..........((((((....)))))).......................--- ( -21.80, z-score = -2.37, R) >droMoj3.scaffold_6540 26910058 92 - 34148556 ---ACUUCGGAAUUACU-CGGCCGAAUUUCA----AAACU--------------AUGAAAUUGGUAAUUAAUCAAAGUCCUUUUCAGGACUACUAUUACAUAUUUCUUAGAGAA--- ---.(....).....((-(.(((.(((((((----.....--------------.))))))))))..........((((((....))))))..................)))..--- ( -19.20, z-score = -2.35, R) >consensus _UAUUUUGUACAUGUUCGCAAUCGAAAUUUU____AGAUUUAU_______AGAUCUGAAAUUU__AUUUAAACAA_GCCCUUUUCAGGGCUACAACGUUAUGUUGC_AAGAGAA___ ............................................................................(((((....)))))........................... ( -6.03 = -5.86 + -0.16)

| Location | 21,527,405 – 21,527,510 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 63.10 |
| Shannon entropy | 0.75309 |
| G+C content | 0.31909 |
| Mean single sequence MFE | -21.42 |
| Consensus MFE | -6.57 |
| Energy contribution | -6.55 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.80 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.981098 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 21527405 105 - 23011544 ---UUCUCUU-GCAACGGAACGUUGUAGCCCUGAAAAGGAC-UUGUUUAAAC--AAAUUUCAGAUCUAUAAAACAUAAAUCU----AAAAUUUUAAUUGCGAACAUGUACCAAAUA- ---.....((-((((((...)))))))).(((....)))((-.(((((.((.--((((((.((((.(((.....))).))))----.))))))...))..))))).))........- ( -17.10, z-score = -1.49, R) >droSim1.chrU 3629637 105 + 15797150 ---UUCUCUU-GCAAAAGAACGUUGUAGCCCUGAAAAGGAC-UUGUAUAAAC--AAAUUUCAGAUCUAUAAAUCAUAAAUCU----AAAUUUUUGAUUGCGAACAUGUACCAAAUA- ---.....((-((((.......((((((..((((((.....-((((....))--)).))))))..)))))).(((.((((..----..)))).)))))))))..............- ( -15.70, z-score = -0.98, R) >droSec1.super_402 1611 105 - 12115 ---UUCUCUU-GCAACAGAACGUUGUAGCCCUGAAAAGGAC-UUGUAUAAAC--AAAUUUCAGAUCUAUAAAUCAUAAAUCU----AAAGUUUUGAUUGCGAACAUGUACCAAAUA- ---.....((-(((((((((((((..((.(((....))).)-)......)))--.......((((.(((.....))).))))----...)))))).))))))..............- ( -19.40, z-score = -1.82, R) >droYak2.chr2R_random 60132 98 + 67238 ---UUCUCUU-GCAACGAAUCGUUGUAGCCCUGAAAAGGGC-UUGUUUAAAC--AAAUUUCAGAUGU-------UGAAAUCU----AAAAUUUUGAUUGCGAACAUGUACUAACUA- ---......(-((((((...)))))))(((((....)))))-.(((((.(((--((((((.((((..-------....))))----.))).)))).))..)))))...........- ( -23.60, z-score = -2.91, R) >droEre2.scaffold_3221 10478 98 + 23538 ---UUCUCUU-GCAAGUGAAUGUUGUAGCCCUGAAAAGGGC-UUGAUUAGAC--AAAUUUCAGAUCU-------AGAAAUCU----AAAAUUUUGAUUGCGAACAUGUACCAACUA- ---.....((-((((.....((((..((((((....)))))-)......)))--)....(((((..(-------((....))----)....)))))))))))..............- ( -22.20, z-score = -1.85, R) >droAna3.scaffold_13030 44304 99 + 47400 ---UUCUCUUUAAAACAUAU-CUUGUAGCCCUGAAAAGGGC-UUGUUUAAUU--UAAUUUUAGAUUU-------CGAAAUCU----AAAAUUUCGAUUGCAAACAUGUGCAAUUUUA ---.........(((((...-.....((((((....)))))-))))))....--.(((((((((((.-------...)))))----))))))..(((((((......)))))))... ( -25.10, z-score = -4.24, R) >dp4.Unknown_singleton_1517 15560 99 - 24252 ---AUCUCUUUUGAAUAUAGUUUUGUAGCCCUGAAAAGGGC-UUGUUUAAAU--CAAUUUCAGACGG-------AUACAUCU----AAAAUUUCGAAUGCGACCUUGUACACCAAC- ---.((.(.((((((..(((...(((((((((....)))))-((((((.((.--....)).))))))-------.)))).))----)....)))))).).))..............- ( -19.70, z-score = -1.40, R) >droPer1.super_0 3286671 99 - 11822988 ---AUCUCUUUUGAAUAUAGUUUUGUAGCCCUGAAAAGGGC-UUGUUUAAAU--CAAUUUCAGACGG-------AUACAUCU----AAAAUUUCGAAUGCGACCUUGUACACCAAC- ---.((.(.((((((..(((...(((((((((....)))))-((((((.((.--....)).))))))-------.)))).))----)....)))))).).))..............- ( -19.70, z-score = -1.40, R) >droWil1.scaffold_181036 31097 114 - 110856 CGCUUAACGCUGAAACUUAACGC-GCCGACCUGGCUGUGUUUAAGUAAAAUUUUAGAUUUUCGUAUUCGAUAUCUAAAAUUUUUUCAAUAUUACGAUUGCAAUAAAGUAUUAAUU-- .((((...((....(((((((((-(((.....))).))).))))))((((((((((((..(((....))).))))))))))))...............))....)))).......-- ( -24.80, z-score = -2.37, R) >droVir3.scaffold_13047 1640872 100 - 19223366 ---UUCUCUUAGAAAAUCGUAUUUGUAGCCCUUAAAAGGGCUUUGUAGGAAAACAAAUUUCAUUAUU-------AGAAAUUU----GCAUAUGCUGACA-AGCAGUUAGCAAUUU-- ---.............((.(((....((((((....))))))..))).))...((((((((......-------.)))))))----)....(((((((.-....)))))))....-- ( -26.90, z-score = -3.33, R) >droMoj3.scaffold_6540 26910058 92 + 34148556 ---UUCUCUAAGAAAUAUGUAAUAGUAGUCCUGAAAAGGACUUUGAUUAAUUACCAAUUUCAU--------------AGUUU----UGAAAUUCGGCCG-AGUAAUUCCGAAGU--- ---(((.(((.(((((..((((((((((((((....))))))...))).)))))..))))).)--------------))...----.))).(((((..(-(....)))))))..--- ( -21.40, z-score = -2.83, R) >consensus ___UUCUCUU_GCAACAUAACGUUGUAGCCCUGAAAAGGGC_UUGUUUAAAC__AAAUUUCAGAUCU_______AUAAAUCU____AAAAUUUCGAUUGCGAACAUGUACAAAAUA_ ...........................(((((....)))))............................................................................ ( -6.57 = -6.55 + -0.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:56:38 2011