| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,522,584 – 21,522,674 |
| Length | 90 |
| Max. P | 0.998597 |

| Location | 21,522,584 – 21,522,674 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 56.59 |
| Shannon entropy | 0.83482 |
| G+C content | 0.30464 |
| Mean single sequence MFE | -18.64 |
| Consensus MFE | -5.65 |
| Energy contribution | -5.65 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.998537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
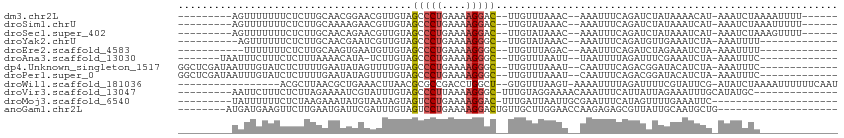
>dm3.chr2L 21522584 90 + 23011544 ------AAAAUUUUAGAUUU-AUGUUUUAUAGAUCUGAAAUUU--GUUUAAACAA--GUCCUUUUCAGGGCUACAACGUUCCGUUGCAAGAGAAAAAAACU--------- ------.(((((((((((((-(((...))))))))))))))))--((((.....(--(((((....)))))).(((((...)))))..........)))).--------- ( -22.60, z-score = -3.49, R) >droSim1.chrU 36846 90 + 15797150 ------AAAAAUUUAGAUUU-AUGAUUUAUAGAUCUGAAAUUU--GUUUAUACAA--GUCCUUUUCAGGGCUACAACGUUCUUUUGCAAGAGAAAAAAACU--------- ------.....(((((((((-(((...))))))))))))....--((((.....(--(((((....))))))......((((((....))))))..)))).--------- ( -18.60, z-score = -2.25, R) >droSec1.super_402 2902 90 + 12115 ------AAAACUUUAGAUUU-AUGAUUUAUAGAUCUGAAAUUU--GUUUAUACAA--GUCCUUUUCAGGGCUACAACGUUCUGUUGCAAGAGAAAAAAACU--------- ------.(((.(((((((((-(((...)))))))))))).)))--((((.....(--(((((....)))))).((((.....))))..........)))).--------- ( -17.70, z-score = -1.60, R) >droYak2.chrU 21897419 82 + 28119190 -------------AAAAUUU-UAGAUUUCAACAUCUGAAAUUU--GUUUAUACAA--GCCCUUUUCAGGGCUACAACGAUUCGUUGCAAGAGAAAAAACU---------- -------------.((((((-(((((......)))))))))))--((((.....(--(((((....)))))).(((((...))))).........)))).---------- ( -20.90, z-score = -3.89, R) >droEre2.scaffold_4583 7495 81 - 11978 -------------AAAAUUU-UAGAUUUCUAGAUCUGAAAUUU--GUCUAAACAA--GCCCUUUUCAGGGCUACAACAUUCACUUGCAAGAGAAAAAAA----------- -------------.((((((-((((((....))))))))))))--.........(--(((((....))))))......(((.((....)).))).....----------- ( -18.90, z-score = -2.85, R) >droAna3.scaffold_13030 44327 84 - 47400 -------------GAAAUUU-UAGAUUUCGAAAUCUAAAAUUA--AAUUAAACAA--GCCCUUUUCAGGGCUACAAGA-UAUGUUUUAAAGAGAAAGAAAUUA------- -------------..(((((-((((((....))))))))))).--.........(--(((((....))))))......-........................------- ( -16.90, z-score = -2.74, R) >dp4.Unknown_singleton_1517 10427 92 + 24252 -------------GAAAUUU-UAGAUGUAUCCGUCUGAAAUUG--AUUUAAACAA--GCCCUUUUCAGGGCUACAAAACUAUAUUCAAAAGAGAUACAAAUUAUCGAGCC -------------..(((((-((((((....))))))))))).--.........(--(((((....))))))....................((((.....))))..... ( -19.80, z-score = -2.98, R) >droPer1.super_0 3279948 92 + 11822988 -------------GAAAUUU-UAGAUGUAUCCGUCUGAAAUUG--AUUUAAACAA--GCCCUUUUCAGGGCUACAAAACUAUAUUCAAAAGAGAUACAAAUUAUCGAGCC -------------..(((((-((((((....))))))))))).--.........(--(((((....))))))....................((((.....))))..... ( -19.80, z-score = -2.98, R) >droWil1.scaffold_181036 31123 89 + 110856 AUUGAAAAAAUUUUAGAUAU-CGAAUACGAAAAUCUAAAAUUUU-ACUUAAACAC--AGCCAGGUCGGCGCGUUAAGUUUCAGCGUUAAGCGU----------------- ..((((((((((((((((.(-((....)))..))))))))))))-((((((....--.(((.....)))...))))))))))((.....))..----------------- ( -22.00, z-score = -2.86, R) >droVir3.scaffold_13047 1623303 86 + 19223366 --------------GCAUAUGCAAAUUUCUAAUAAUGAAAUUUGUUUUCCUACAAA-GCCCUUUUAAGGGCUACAAAUACGAUUUUCUAAGAGAAAGAAUU--------- --------------......(((((((((.......)))))))))..........(-(((((....))))))..........(((((.....)))))....--------- ( -18.50, z-score = -2.77, R) >droMoj3.scaffold_6540 26892971 79 - 34148556 ---------------------GAAUUUCAAAACUAUGAAAUUCGCAAUUAAUCAAA-GUCCUUUUCAGGACUACUAUUACAUAUUUCUUAGAGAAAAAAUA--------- ---------------------((((((((......))))))))............(-(((((....))))))...........((((.....)))).....--------- ( -15.30, z-score = -3.66, R) >anoGam1.chr2L 35944003 82 - 48795086 --------------------CAGCAUUGCAAUAACGCUCUCUUGGUUCCAAGCAACAGUCCUUUUCAGGACUACAAAUCGAAUCAUUCAAGAACUUCAUCAU-------- --------------------.......((......))(((..((((((........((((((....)))))).......))))))....)))..........-------- ( -12.66, z-score = -1.27, R) >consensus _____________UAAAUUU_UAGAUUUAUAAAUCUGAAAUUU__AUUUAAACAA__GCCCUUUUCAGGGCUACAACAUUAUGUUGCAAAGAAAAAAAACU_________ .........................................................(((((....)))))....................................... ( -5.65 = -5.65 + -0.00)

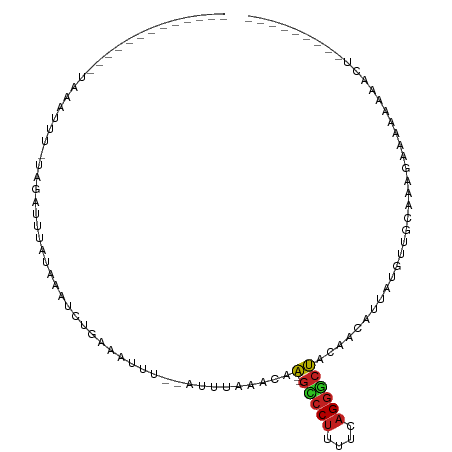
| Location | 21,522,584 – 21,522,674 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 56.59 |
| Shannon entropy | 0.83482 |
| G+C content | 0.30464 |
| Mean single sequence MFE | -20.14 |
| Consensus MFE | -5.92 |
| Energy contribution | -6.18 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.998597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 21522584 90 - 23011544 ---------AGUUUUUUUCUCUUGCAACGGAACGUUGUAGCCCUGAAAAGGAC--UUGUUUAAAC--AAAUUUCAGAUCUAUAAAACAU-AAAUCUAAAAUUUU------ ---------((((((((((..((((((((...))))))))....)))))))))--).........--((((((.((((.(((.....))-).)))).)))))).------ ( -18.30, z-score = -2.46, R) >droSim1.chrU 36846 90 - 15797150 ---------AGUUUUUUUCUCUUGCAAAAGAACGUUGUAGCCCUGAAAAGGAC--UUGUAUAAAC--AAAUUUCAGAUCUAUAAAUCAU-AAAUCUAAAUUUUU------ ---------.(((((((((....).)))))))).((((((..((((((.....--((((....))--)).))))))..)))))).....-..............------ ( -13.90, z-score = -1.20, R) >droSec1.super_402 2902 90 - 12115 ---------AGUUUUUUUCUCUUGCAACAGAACGUUGUAGCCCUGAAAAGGAC--UUGUAUAAAC--AAAUUUCAGAUCUAUAAAUCAU-AAAUCUAAAGUUUU------ ---------((((((((((..(((((((.....)))))))....)))))))))--).........--((((((.((((.(((.....))-).)))).)))))).------ ( -17.50, z-score = -2.23, R) >droYak2.chrU 21897419 82 - 28119190 ----------AGUUUUUUCUCUUGCAACGAAUCGUUGUAGCCCUGAAAAGGGC--UUGUAUAAAC--AAAUUUCAGAUGUUGAAAUCUA-AAAUUUU------------- ----------.((((...(...(((((((...)))))))(((((....)))))--..)...))))--((((((.((((......)))).-)))))).------------- ( -22.40, z-score = -3.82, R) >droEre2.scaffold_4583 7495 81 + 11978 -----------UUUUUUUCUCUUGCAAGUGAAUGUUGUAGCCCUGAAAAGGGC--UUGUUUAGAC--AAAUUUCAGAUCUAGAAAUCUA-AAAUUUU------------- -----------.....(((.((....)).)))((((..((((((....)))))--)......)))--)(((((.((((......)))).-)))))..------------- ( -16.90, z-score = -1.15, R) >droAna3.scaffold_13030 44327 84 + 47400 -------UAAUUUCUUUCUCUUUAAAACAUA-UCUUGUAGCCCUGAAAAGGGC--UUGUUUAAUU--UAAUUUUAGAUUUCGAAAUCUA-AAAUUUC------------- -------.................(((((..-......((((((....)))))--))))))....--.(((((((((((....))))))-)))))..------------- ( -19.40, z-score = -4.06, R) >dp4.Unknown_singleton_1517 10427 92 - 24252 GGCUCGAUAAUUUGUAUCUCUUUUGAAUAUAGUUUUGUAGCCCUGAAAAGGGC--UUGUUUAAAU--CAAUUUCAGACGGAUACAUCUA-AAAUUUC------------- ............(((((((..((((((....((((...((((((....)))))--).....))))--....)))))).)))))))....-.......------------- ( -23.50, z-score = -3.17, R) >droPer1.super_0 3279948 92 - 11822988 GGCUCGAUAAUUUGUAUCUCUUUUGAAUAUAGUUUUGUAGCCCUGAAAAGGGC--UUGUUUAAAU--CAAUUUCAGACGGAUACAUCUA-AAAUUUC------------- ............(((((((..((((((....((((...((((((....)))))--).....))))--....)))))).)))))))....-.......------------- ( -23.50, z-score = -3.17, R) >droWil1.scaffold_181036 31123 89 - 110856 -----------------ACGCUUAACGCUGAAACUUAACGCGCCGACCUGGCU--GUGUUUAAGU-AAAAUUUUAGAUUUUCGUAUUCG-AUAUCUAAAAUUUUUUCAAU -----------------..((.....))((((((((((((((((.....))).--))).))))))-((((((((((((..(((....))-).)))))))))))))))).. ( -24.60, z-score = -4.16, R) >droVir3.scaffold_13047 1623303 86 - 19223366 ---------AAUUCUUUCUCUUAGAAAAUCGUAUUUGUAGCCCUUAAAAGGGC-UUUGUAGGAAAACAAAUUUCAUUAUUAGAAAUUUGCAUAUGC-------------- ---------..((((.......))))..((.(((....((((((....)))))-)..))).))...((((((((.......)))))))).......-------------- ( -19.20, z-score = -2.42, R) >droMoj3.scaffold_6540 26892971 79 + 34148556 ---------UAUUUUUUCUCUAAGAAAUAUGUAAUAGUAGUCCUGAAAAGGAC-UUUGAUUAAUUGCGAAUUUCAUAGUUUUGAAAUUC--------------------- ---------.....((((.(((.(((((.(((((((((((((((....)))))-)...))).)))))).))))).)))....))))...--------------------- ( -20.10, z-score = -3.80, R) >anoGam1.chr2L 35944003 82 + 48795086 --------AUGAUGAAGUUCUUGAAUGAUUCGAUUUGUAGUCCUGAAAAGGACUGUUGCUUGGAACCAAGAGAGCGUUAUUGCAAUGCUG-------------------- --------..(((((.((((((....(.(((((.(..(((((((....)))))))..).))))).)...)))))).))))).........-------------------- ( -22.40, z-score = -2.70, R) >consensus _________AGUUUUUUUCCCUUGAAACAGAAAGUUGUAGCCCUGAAAAGGGC__UUGUUUAAAC__AAAUUUCAGAUCUAUAAAUCUA_AAAUUUA_____________ .......................................(((((....)))))......................................................... ( -5.92 = -6.18 + 0.26)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:56:37 2011