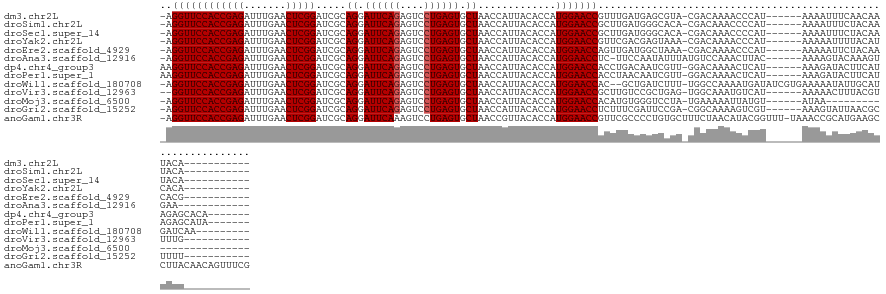
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,009,493 – 2,009,609 |
| Length | 116 |
| Max. P | 0.994367 |

| Location | 2,009,493 – 2,009,609 |
|---|---|
| Length | 116 |
| Sequences | 13 |
| Columns | 135 |
| Reading direction | forward |
| Mean pairwise identity | 76.63 |
| Shannon entropy | 0.50137 |
| G+C content | 0.44854 |
| Mean single sequence MFE | -31.53 |
| Consensus MFE | -25.48 |
| Energy contribution | -25.48 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.62 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.779514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
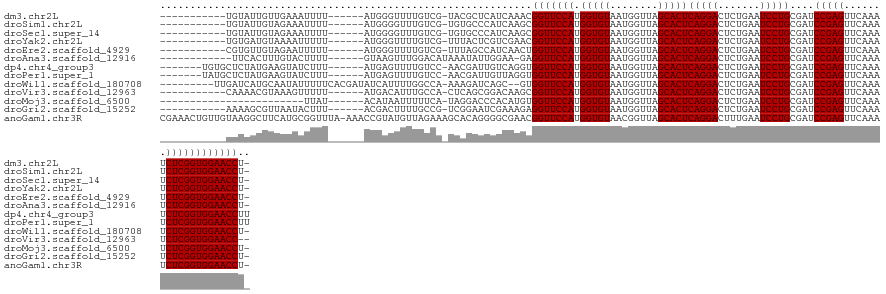
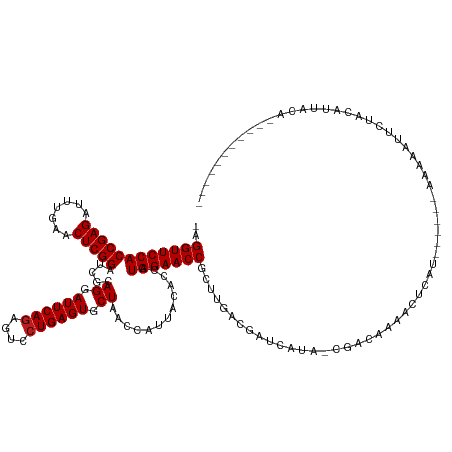
>dm3.chr2L 2009493 116 + 23011544 -----------UGUAUUGUUGAAAUUUU------AUGGGUUUUGUCG-UACGCUCAUCAAACGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCU- -----------.............(((.------((((((.(.....-.).)))))).))).(((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))).- ( -29.10, z-score = -0.27, R) >droSim1.chr2L 1979155 116 + 22036055 -----------UGUAUUGUAGAAAUUUU------AUGGGGUUUGUCG-UGUGCCCAUCAAGCGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCU- -----------.................------...((((......-...)))).......(((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))).- ( -30.90, z-score = -0.13, R) >droSec1.super_14 1956320 116 + 2068291 -----------UGUAUUGUAGAAAUUUU------AUGGGGUUUGUCG-UGUGCCCAUCAAGCGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCU- -----------.................------...((((......-...)))).......(((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))).- ( -30.90, z-score = -0.13, R) >droYak2.chr2L 1994465 116 + 22324452 -----------UGUGAUGUAAAAUUUUU------AUGGGUUUUGUCG-UUUACUCGUCGAACGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCU- -----------.((((((((((((((..------..)))))))).))-.)))).........(((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))).- ( -30.40, z-score = -0.60, R) >droEre2.scaffold_4929 2055467 116 + 26641161 -----------CGUGUUGUAGAAUUUUU------AUGGGUUUUGUCG-UUUAGCCAUCAACUGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCU- -----------...(((((((.((((..------..((((((((..(-(((((((((..((((......))))...)))))))).)).)))))))).)))).)))))))(((((.......)))))........- ( -32.30, z-score = -1.11, R) >droAna3.scaffold_12916 9725169 115 - 16180835 ------------UUCACUUUGUACUUUU------GUAAGUUUGGACAUAAAUAUUGGAA-GAGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCU- ------------...........(((..------(...(((((....)))))..)..))-)((((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))))- ( -28.90, z-score = -0.33, R) >dp4.chr4_group3 8788677 121 + 11692001 -------UGUGCUCUAUGAAGUAUCUUU------AUGAGUUUUGUCC-AACGAUUGUCAGGUGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCUU -------...((((.((((((...))))------))))))....(((-(..(((((.((((.(..(((.((((((........)))).)))))..).....)))))))))((((.......)))).))))..... ( -33.10, z-score = -0.73, R) >droPer1.super_1 10240448 121 + 10282868 -------UAUGCUCUAUGAAGUAUCUUU------AUGAGUUUUGUCC-AACGAUUGUUAGGUGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCUU -------...((((.((((((...))))------)))))).....((-(((....))).)).(((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))).. ( -32.10, z-score = -0.78, R) >droWil1.scaffold_180708 11650997 122 - 12563649 ---------UUGAUCAUGCAAUAUUUUUCACGAUAUCAUUUUGGCCA-AAAGAUCAGC--GUGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCU- ---------((((((.....(((((......)))))..((((....)-))))))))).--..(((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))).- ( -29.60, z-score = 0.14, R) >droVir3.scaffold_12963 7605753 115 - 20206255 -----------CAAAACGUAAAGUUUUU------AUGACAUUUGCCA-CUCAGCGGACAAGCGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACC-- -----------......((((((((...------..))).)))))..-....((......))(((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))-- ( -30.00, z-score = -0.55, R) >droMoj3.scaffold_6500 29329693 103 + 32352404 ------------------------UUAU------ACAUAAUUUUUCA-UAGGACCCACAUGUGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCU- ------------------------...(------((((.......((-(.(((.(((....))).)))))))))))..((((..(((.(((((.......))))).)..(((((.......))))))).)))).- ( -25.81, z-score = -0.34, R) >droGri2.scaffold_15252 164294 116 + 17193109 -----------AAAAGCGUUAAUACUUU------ACGACUUUUGCCG-UCGGAAUCGAAAGAGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCU- -----------((((((((.........------))).)))))((((-(.((((((......))))))))))).....((((..(((.(((((.......))))).)..(((((.......))))))).)))).- ( -33.10, z-score = -1.16, R) >anoGam1.chr3R 22017966 133 - 53272125 CGAAACUGUUGUAAGGCUUCAUGCGGUUUA-AAACCGUAUGUUAGAAAGCACAGGGGCGAACGGUUCCAUGGUGUAACGGUUAGCACUCAGGACUUUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCU- ((...((((.((....((.((((((((...-..))))))))..))...))))))...))...(((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))).- ( -43.70, z-score = -2.07, R) >consensus ___________UGUAAUGUAGAAUUUUU______AUGAGUUUUGUCG_UACGAUCGUCAAGCGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCU_ ..............................................................(((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))).. (-25.48 = -25.48 + 0.00)
| Location | 2,009,493 – 2,009,609 |
|---|---|
| Length | 116 |
| Sequences | 13 |
| Columns | 135 |
| Reading direction | reverse |
| Mean pairwise identity | 76.63 |
| Shannon entropy | 0.50137 |
| G+C content | 0.44854 |
| Mean single sequence MFE | -30.83 |
| Consensus MFE | -26.45 |
| Energy contribution | -26.53 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.994367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
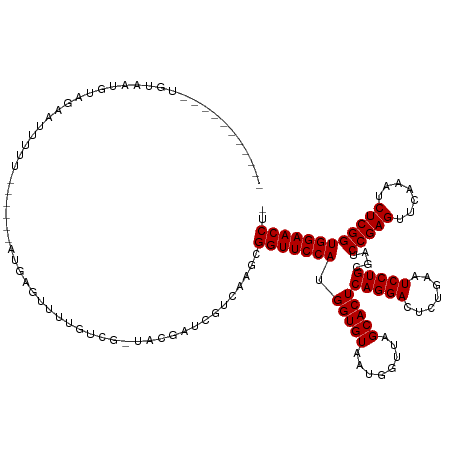
>dm3.chr2L 2009493 116 - 23011544 -AGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCGUUUGAUGAGCGUA-CGACAAAACCCAU------AAAAUUUCAACAAUACA----------- -.((((((((((((.......))))).....((.((((((....)))))).)).............)))))))(((((.((((....-.............------.....)))).))).)).----------- ( -28.05, z-score = -1.75, R) >droSim1.chr2L 1979155 116 - 22036055 -AGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCGCUUGAUGGGCACA-CGACAAACCCCAU------AAAAUUUCUACAAUACA----------- -.((((((((((((.......))))).....((.((((((....)))))).)).............))))))).....(((((....-........)))))------.................----------- ( -31.30, z-score = -2.44, R) >droSec1.super_14 1956320 116 - 2068291 -AGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCGCUUGAUGGGCACA-CGACAAACCCCAU------AAAAUUUCUACAAUACA----------- -.((((((((((((.......))))).....((.((((((....)))))).)).............))))))).....(((((....-........)))))------.................----------- ( -31.30, z-score = -2.44, R) >droYak2.chr2L 1994465 116 - 22324452 -AGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCGUUCGACGAGUAAA-CGACAAAACCCAU------AAAAAUUUUACAUCACA----------- -.((((((((((((.......))))).....((.((((((....)))))).)).............))))))).......(((((((-.............------......))))).))...----------- ( -27.91, z-score = -1.94, R) >droEre2.scaffold_4929 2055467 116 - 26641161 -AGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCAGUUGAUGGCUAAA-CGACAAAACCCAU------AAAAAUUCUACAACACG----------- -.((((((((((((.......))))).....((.((((((....)))))).)).............))))))).((((((((.....-.........))))------..........))))...----------- ( -28.94, z-score = -2.21, R) >droAna3.scaffold_12916 9725169 115 + 16180835 -AGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCUC-UUCCAAUAUUUAUGUCCAAACUUAC------AAAAGUACAAAGUGAA------------ -(((((((((((((.......))))).....((.((((((....)))))).)).............)))))))).-......(((((.(((........))------).))))).........------------ ( -28.20, z-score = -1.73, R) >dp4.chr4_group3 8788677 121 - 11692001 AAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCACCUGACAAUCGUU-GGACAAAACUCAU------AAAGAUACUUCAUAGAGCACA------- ..((((((((((((.......))))).....((.((((((....)))))).)).............)))))))..(..((....)).-.)......(((((------.(((...))).)).)))....------- ( -30.30, z-score = -1.62, R) >droPer1.super_1 10240448 121 - 10282868 AAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCACCUAACAAUCGUU-GGACAAAACUCAU------AAAGAUACUUCAUAGAGCAUA------- ..((((((((((((.......))))).....((.((((((....)))))).)).............)))))))..(((((....)))-))......(((((------.(((...))).)).)))....------- ( -30.80, z-score = -2.24, R) >droWil1.scaffold_180708 11650997 122 + 12563649 -AGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCAC--GCUGAUCUUU-UGGCCAAAAUGAUAUCGUGAAAAAUAUUGCAUGAUCAA--------- -.((((((((((((.......))))).....((.((((((....)))))).)).............)))))))..--((..(....)-..)).....((((...(..(......)..)...)))).--------- ( -33.80, z-score = -1.86, R) >droVir3.scaffold_12963 7605753 115 + 20206255 --GGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCGCUUGUCCGCUGAG-UGGCAAAUGUCAU------AAAAACUUUACGUUUUG----------- --((((((((((((.......))))).....((.((((((....)))))).)).............)))))))((......))...(-((((....)))))------.(((((.....))))).----------- ( -33.00, z-score = -1.81, R) >droMoj3.scaffold_6500 29329693 103 - 32352404 -AGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCACAUGUGGGUCCUA-UGAAAAAUUAUGU------AUAA------------------------ -.((((((((((((.......))))).....((.((((((....)))))).)).............)))))))(((((....((...-.)).....)))))------....------------------------ ( -28.90, z-score = -1.49, R) >droGri2.scaffold_15252 164294 116 - 17193109 -AGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCUCUUUCGAUUCCGA-CGGCAAAAGUCGU------AAAGUAUUAACGCUUUU----------- -(((((((((((((.......))))).....((.((((((....)))))).)).............))))))))............(-((((....)))))------(((((......))))).----------- ( -32.90, z-score = -1.95, R) >anoGam1.chr3R 22017966 133 + 53272125 -AGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAAAGUCCUGAGUGCUAACCGUUACACCAUGGAACCGUUCGCCCCUGUGCUUUCUAACAUACGGUUU-UAAACCGCAUGAAGCCUUACAACAGUUUCG -.((((((((((((.......)))))....((((((.....))))))...................)))))))........((((((((.....(((.((((..-...)))).)))))))......))))..... ( -35.40, z-score = -1.63, R) >consensus _AGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCGCUUGACGAUCAUA_CGACAAAACUCAU______AAAAAUUCUACAUUACA___________ ..((((((((((((.......))))).....((.((((((....)))))).)).............))))))).............................................................. (-26.45 = -26.53 + 0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:50 2011