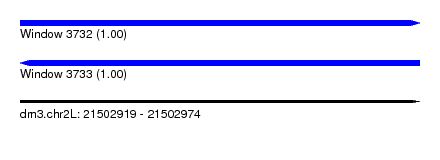
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,502,919 – 21,502,974 |
| Length | 55 |
| Max. P | 0.999888 |

| Location | 21,502,919 – 21,502,974 |
|---|---|
| Length | 55 |
| Sequences | 11 |
| Columns | 70 |
| Reading direction | forward |
| Mean pairwise identity | 58.69 |
| Shannon entropy | 0.83541 |
| G+C content | 0.47896 |
| Mean single sequence MFE | -19.95 |
| Consensus MFE | -12.49 |
| Energy contribution | -13.75 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.72 |
| SVM RNA-class probability | 0.999888 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
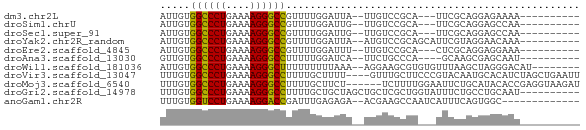
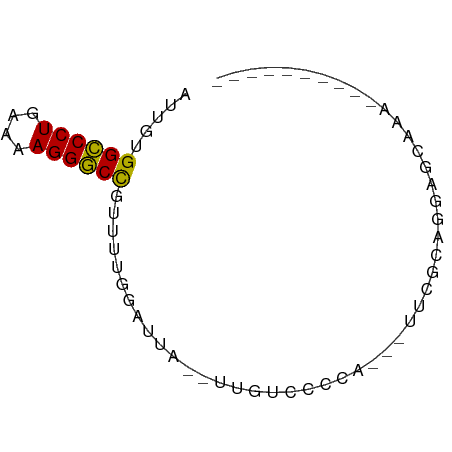
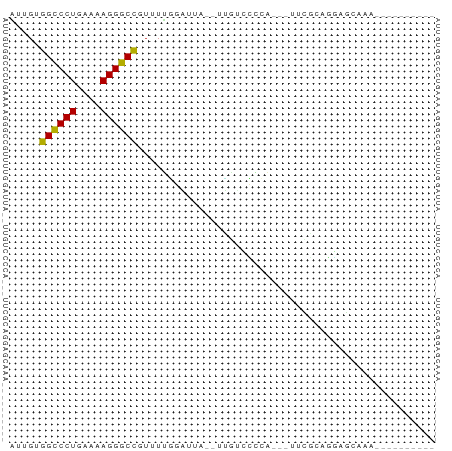
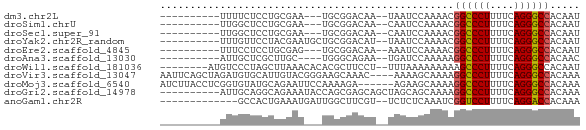
>dm3.chr2L 21502919 55 + 23011544 AUUGUGGCCCUGAAAAGGGCCGUUUUGGAUUA--UUGUCCGCA---UUCGCAGGAGAAAA---------- .((((((((((....)))))).....((((..--..))))...---...)))).......---------- ( -20.10, z-score = -2.58, R) >droSim1.chrU 5518301 55 - 15797150 AUUGUGGCCCUGAAAAGGGCCGUUUUGGAUUG--UUGUCCGCA---UUCGCAGGAGCCAA---------- .....((((((....))))))(((((((((..--..))))((.---...)).)))))...---------- ( -21.60, z-score = -2.34, R) >droSec1.super_91 112088 55 + 125313 AUUGUGGCCCUGAAAAGGGCCGUUUUGGAUUG--UUGUCCGCA---UUCGCAGGAGCCAA---------- .....((((((....))))))(((((((((..--..))))((.---...)).)))))...---------- ( -21.60, z-score = -2.34, R) >droYak2.chr2R_random 60460 58 - 67238 AUUGUGGCCCUGAAAAGGGCCGUUUUGGAUUA--AUGUCCGCAGCAUUCGUAGGAACAAA---------- .((((((((((....))))))(((.(((((..--..))))).)))..........)))).---------- ( -23.20, z-score = -3.62, R) >droEre2.scaffold_4845 19574780 55 - 22589142 AUUGUGGCCCUGAAAAGGGCCGUUUUGGAUUU--UUGUCCGCA---CUCGCAGGAGGAAA---------- ..(((((((((....)))))).....((((..--..)))))))---(((....)))....---------- ( -21.40, z-score = -2.38, R) >droAna3.scaffold_13030 44769 54 - 47400 GUUGUGGCCCUGAAAAGGGCCUUUUUGGAUCA--UUCUGCCCA----GCAAGCGAGCAAU---------- ((((.((((((....)))))).....(((...--.)))...))----))...........---------- ( -17.50, z-score = -0.99, R) >droWil1.scaffold_181036 1021 60 + 110856 AUUGUGGCCCUGAAAAGGGCUUUUUUUUUAAA--AGGAAGCGUGUGUUUAAGCUAGGGACAU-------- .((.(((((((....)))((((((((.....)--)))))))..........)))).))....-------- ( -14.80, z-score = -0.85, R) >droVir3.scaffold_13047 1603366 66 + 19223366 UUUGUGGCCCUGAAAAGGGCCUUUUGCUUUU----GUUUGCUUCCCGUACAAUGCACAUCUAGCUGAAUU .....((((((....))))))(((.(((...----((.(((............))).))..))).))).. ( -18.20, z-score = -1.84, R) >droMoj3.scaffold_6540 26857080 64 - 34148556 UUUGUGGCCCUGAAAAGGGCCUUUUGCUUCU------UCUUUUGGAAUUCUGCAUACACCGAGGUAAGAU .....((((((....))))))(((((((((.------.....((.(....).))......))))))))). ( -20.80, z-score = -2.43, R) >droGri2.scaffold_14978 520668 60 + 1124632 UUUGUGGCCCUGAAAAGGGCCUUUUGCUGCUAGCUGCUCGCUGGUAUUUCUGCCUGCAAU---------- .((((((((((....))))))......(((((((.....))))))).........)))).---------- ( -23.60, z-score = -2.79, R) >anoGam1.chr2R 47876183 55 + 62725911 UUUGUGGUCCUGAAAAGGACCGAUUUGAGAGA--ACGAAGCCAAUCAUUUCAGUGGC------------- (((((((((((....))))))...((....))--)))))((((..........))))------------- ( -16.60, z-score = -2.43, R) >consensus AUUGUGGCCCUGAAAAGGGCCGUUUUGGAUUA__UUGUCCCCA___UUCGCAGGAGCAAA__________ .....((((((....))))))................................................. (-12.49 = -13.75 + 1.25)
| Location | 21,502,919 – 21,502,974 |
|---|---|
| Length | 55 |
| Sequences | 11 |
| Columns | 70 |
| Reading direction | reverse |
| Mean pairwise identity | 58.69 |
| Shannon entropy | 0.83541 |
| G+C content | 0.47896 |
| Mean single sequence MFE | -15.70 |
| Consensus MFE | -11.22 |
| Energy contribution | -11.15 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.13 |
| SVM RNA-class probability | 0.999648 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
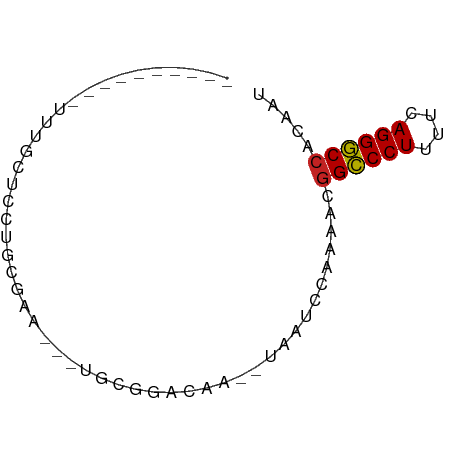
>dm3.chr2L 21502919 55 - 23011544 ----------UUUUCUCCUGCGAA---UGCGGACAA--UAAUCCAAAACGGCCCUUUUCAGGGCCACAAU ----------.......((((...---.))))....--...........((((((....))))))..... ( -15.70, z-score = -2.52, R) >droSim1.chrU 5518301 55 + 15797150 ----------UUGGCUCCUGCGAA---UGCGGACAA--CAAUCCAAAACGGCCCUUUUCAGGGCCACAAU ----------(((..(((.((...---.)))))...--)))........((((((....))))))..... ( -16.30, z-score = -1.62, R) >droSec1.super_91 112088 55 - 125313 ----------UUGGCUCCUGCGAA---UGCGGACAA--CAAUCCAAAACGGCCCUUUUCAGGGCCACAAU ----------(((..(((.((...---.)))))...--)))........((((((....))))))..... ( -16.30, z-score = -1.62, R) >droYak2.chr2R_random 60460 58 + 67238 ----------UUUGUUCCUACGAAUGCUGCGGACAU--UAAUCCAAAACGGCCCUUUUCAGGGCCACAAU ----------(((((....)))))......(((...--...))).....((((((....))))))..... ( -15.40, z-score = -2.01, R) >droEre2.scaffold_4845 19574780 55 + 22589142 ----------UUUCCUCCUGCGAG---UGCGGACAA--AAAUCCAAAACGGCCCUUUUCAGGGCCACAAU ----------.......((((...---.))))....--...........((((((....))))))..... ( -15.70, z-score = -1.81, R) >droAna3.scaffold_13030 44769 54 + 47400 ----------AUUGCUCGCUUGC----UGGGCAGAA--UGAUCCAAAAAGGCCCUUUUCAGGGCCACAAC ----------.(((((((.....----)))))))..--...........((((((....))))))..... ( -18.10, z-score = -1.59, R) >droWil1.scaffold_181036 1021 60 - 110856 --------AUGUCCCUAGCUUAAACACACGCUUCCU--UUUAAAAAAAAAGCCCUUUUCAGGGCCACAAU --------.(((....(((..........)))....--............(((((....))))).))).. ( -8.00, z-score = -0.71, R) >droVir3.scaffold_13047 1603366 66 - 19223366 AAUUCAGCUAGAUGUGCAUUGUACGGGAAGCAAAC----AAAAGCAAAAGGCCCUUUUCAGGGCCACAAA ......(((...(((((...)))))...)))....----..........((((((....))))))..... ( -18.30, z-score = -1.74, R) >droMoj3.scaffold_6540 26857080 64 + 34148556 AUCUUACCUCGGUGUAUGCAGAAUUCCAAAAGA------AGAAGCAAAAGGCCCUUUUCAGGGCCACAAA ....(((....)))..(((....(((.....))------)...)))...((((((....))))))..... ( -15.10, z-score = -1.13, R) >droGri2.scaffold_14978 520668 60 - 1124632 ----------AUUGCAGGCAGAAAUACCAGCGAGCAGCUAGCAGCAAAAGGCCCUUUUCAGGGCCACAAA ----------.((((.((.(....).))(((.....)))....))))..((((((....))))))..... ( -20.30, z-score = -2.35, R) >anoGam1.chr2R 47876183 55 - 62725911 -------------GCCACUGAAAUGAUUGGCUUCGU--UCUCUCAAAUCGGUCCUUUUCAGGACCACAAA -------------((((..........)))).....--...........((((((....))))))..... ( -13.50, z-score = -2.00, R) >consensus __________UUUGCUCCUGCGAA___UGCGGACAA__UAAUCCAAAACGGCCCUUUUCAGGGCCACAAU .................................................((((((....))))))..... (-11.22 = -11.15 + -0.07)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:56:32 2011