
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,490,339 – 21,490,412 |
| Length | 73 |
| Max. P | 0.999880 |

| Location | 21,490,339 – 21,490,412 |
|---|---|
| Length | 73 |
| Sequences | 10 |
| Columns | 81 |
| Reading direction | forward |
| Mean pairwise identity | 62.29 |
| Shannon entropy | 0.77945 |
| G+C content | 0.32034 |
| Mean single sequence MFE | -12.63 |
| Consensus MFE | -9.80 |
| Energy contribution | -9.80 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.39 |
| SVM RNA-class probability | 0.999785 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
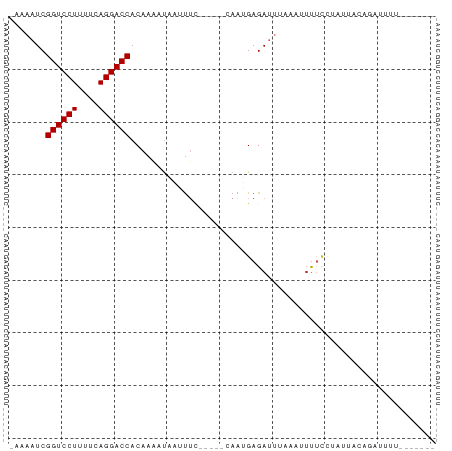
>dm3.chr2L 21490339 73 + 23011544 UAUAAUCGGUCCUUUUCAGGACCACAAACCAGAUUC------AAUGAGA--UAAAAUUUUCUGUUGCCGACUAUUUAUAAC (((((..((((((....))))))............(------(((((((--......)))).))))........))))).. ( -12.90, z-score = -1.71, R) >droYak2.chrU 21895313 73 + 28119190 UACAAUCGGUCCUUUUCAGGACCACAAACCACAUUC------AAUGAGAUUUAAAUUUUUCUUUUGCAGCCUAUUUAUA-- .......((((((....))))))............(------((.((((........))))..))).............-- ( -11.10, z-score = -2.22, R) >droEre2.scaffold_4845 19558368 72 - 22589142 UACAAUCGGUCCUUUUCAGGACCACAAACUACAUUC------AAUGAGAAAUAAAAUUCGCUUUAGCAGCCUUUUUUU--- .......((((((....)))))).............------.....(((......)))((....))...........--- ( -11.40, z-score = -2.06, R) >droAna3.scaffold_13030 42446 68 - 47400 AAAAAUCGGUCCUUUUCAGGACCACAAUGUUAUUUU-----CAAUGAGAUUUGAGUACCUAU---UGAUAUAUAAC----- .......((((((....)))))).(((((.....((-----(((......)))))....)))---)).........----- ( -14.60, z-score = -2.14, R) >dp4.Unknown_singleton_1517 3175 67 + 24252 --AAAUCGGUCCUUUUCAGGACCACAAAAUAAUUUC-----CAAUGAGAUUUUUUAUUUCUGCAUACUCGUACU------- --.....((((((....))))))...(((.((((((-----....)))))).)))...................------- ( -12.10, z-score = -2.50, R) >droPer1.super_0 3272744 67 + 11822988 --AAAUCGGUCCUUUUCAGGACCACAAAAUAAUUUU-----CAAUGAGAUUUUUUAUUUCUGCAUACUCGUACU------- --.....((((((....)))))).............-----....(((((.....)))))..............------- ( -11.60, z-score = -2.35, R) >droWil1.scaffold_181036 3768 64 + 110856 -CAAAUCGGUCCUUUUCAGGACCACAAACUAGUCAUGUCGAUAUUGAGAUUAUUAUUUUCUUAAU---------------- -......((((((....))))))........(((.....)))(((((((.........)))))))---------------- ( -13.20, z-score = -2.27, R) >droVir3.scaffold_13047 1606267 64 + 19223366 --AAAUCGGUCCUUUUCAGGACCACAAAAUAUUUUA---AACAAAGAGAUUGAUAUUUUCCUAUAUACA------------ --.....((((((....))))))..((((((((...---............))))))))..........------------ ( -11.06, z-score = -2.26, R) >droMoj3.scaffold_6540 26915489 72 - 34148556 -CAAAUCGGUCCUUUUCAGGACCACAAUAUUUCUAA-----CAAGGAGAUUUCUAUUUUCGCUUCAACUUUUUUCAAU--- -......((((((....)))))).............-----...((((((....))))))..................--- ( -12.70, z-score = -3.18, R) >droGri2.scaffold_14978 523520 72 + 1124632 -CAAAUCGGUCCUUUUCAGGACCACAAAAUACUUUCA---ACAAAGAGAUUUGAGUUUUCAUAAAUAAUAUUUGAA----- -((((((((((((....))))))........((((..---..)))).)))))).......................----- ( -15.60, z-score = -3.02, R) >consensus _AAAAUCGGUCCUUUUCAGGACCACAAAAUAAUUUC_____CAAUGAGAUUUAAAUUUUCCUAUUACAGAUUUU_______ .......((((((....)))))).......................................................... ( -9.80 = -9.80 + -0.00)


| Location | 21,490,339 – 21,490,412 |
|---|---|
| Length | 73 |
| Sequences | 10 |
| Columns | 81 |
| Reading direction | reverse |
| Mean pairwise identity | 62.29 |
| Shannon entropy | 0.77945 |
| G+C content | 0.32034 |
| Mean single sequence MFE | -16.32 |
| Consensus MFE | -12.50 |
| Energy contribution | -12.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.69 |
| SVM RNA-class probability | 0.999880 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
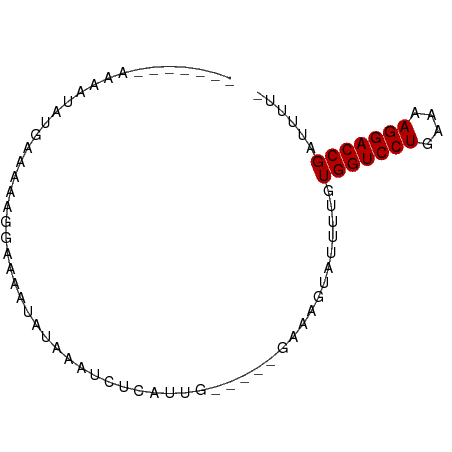
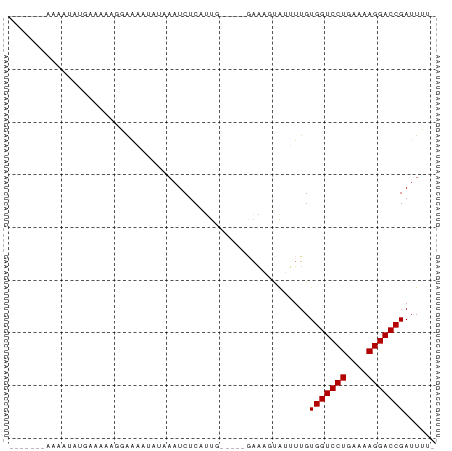
>dm3.chr2L 21490339 73 - 23011544 GUUAUAAAUAGUCGGCAACAGAAAAUUUUA--UCUCAUU------GAAUCUGGUUUGUGGUCCUGAAAAGGACCGAUUAUA .......((((((.(((.((((....((((--......)------)))))))...)))((((((....)))))))))))). ( -20.80, z-score = -3.44, R) >droYak2.chrU 21895313 73 - 28119190 --UAUAAAUAGGCUGCAAAAGAAAAAUUUAAAUCUCAUU------GAAUGUGGUUUGUGGUCCUGAAAAGGACCGAUUGUA --(((((((((((..(((((......)))...((.....------)).))..))))))((((((....))))))..))))) ( -18.80, z-score = -3.47, R) >droEre2.scaffold_4845 19558368 72 + 22589142 ---AAAAAAAGGCUGCUAAAGCGAAUUUUAUUUCUCAUU------GAAUGUAGUUUGUGGUCCUGAAAAGGACCGAUUGUA ---......(((((((..(((.(((......))).).))------....))))))).(((((((....)))))))...... ( -18.50, z-score = -2.89, R) >droAna3.scaffold_13030 42446 68 + 47400 -----GUUAUAUAUCA---AUAGGUACUCAAAUCUCAUUG-----AAAAUAACAUUGUGGUCCUGAAAAGGACCGAUUUUU -----(((((...(((---((((((......)))).))))-----)..)))))....(((((((....)))))))...... ( -20.10, z-score = -4.31, R) >dp4.Unknown_singleton_1517 3175 67 - 24252 -------AGUACGAGUAUGCAGAAAUAAAAAAUCUCAUUG-----GAAAUUAUUUUGUGGUCCUGAAAAGGACCGAUUU-- -------...........((((((.(((....(((....)-----))..)))))))))((((((....)))))).....-- ( -14.10, z-score = -1.94, R) >droPer1.super_0 3272744 67 - 11822988 -------AGUACGAGUAUGCAGAAAUAAAAAAUCUCAUUG-----AAAAUUAUUUUGUGGUCCUGAAAAGGACCGAUUU-- -------...........((((((.(((....((.....)-----)...)))))))))((((((....)))))).....-- ( -14.30, z-score = -2.44, R) >droWil1.scaffold_181036 3768 64 - 110856 ----------------AUUAAGAAAAUAAUAAUCUCAAUAUCGACAUGACUAGUUUGUGGUCCUGAAAAGGACCGAUUUG- ----------------..............((((.........(((.........)))((((((....))))))))))..- ( -13.00, z-score = -1.87, R) >droVir3.scaffold_13047 1606267 64 - 19223366 ------------UGUAUAUAGGAAAAUAUCAAUCUCUUUGUU---UAAAAUAUUUUGUGGUCCUGAAAAGGACCGAUUU-- ------------........(.((((((((((.....)))..---....))))))).)((((((....)))))).....-- ( -13.00, z-score = -1.86, R) >droMoj3.scaffold_6540 26915489 72 + 34148556 ---AUUGAAAAAAGUUGAAGCGAAAAUAGAAAUCUCCUUG-----UUAGAAAUAUUGUGGUCCUGAAAAGGACCGAUUUG- ---..........................(((((......-----.((....))....((((((....))))))))))).- ( -14.20, z-score = -2.39, R) >droGri2.scaffold_14978 523520 72 - 1124632 -----UUCAAAUAUUAUUUAUGAAAACUCAAAUCUCUUUGU---UGAAAGUAUUUUGUGGUCCUGAAAAGGACCGAUUUG- -----.......................((((((.((((..---..))))........((((((....))))))))))))- ( -16.40, z-score = -2.86, R) >consensus _______AAAAUAUGAAAAAGGAAAAUAUAAAUCUCAUUG_____GAAAGUAUUUUGUGGUCCUGAAAAGGACCGAUUUU_ .........................................................(((((((....)))))))...... (-12.50 = -12.50 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:56:29 2011