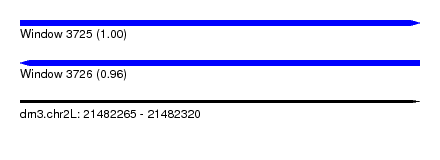
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,482,265 – 21,482,320 |
| Length | 55 |
| Max. P | 0.999415 |

| Location | 21,482,265 – 21,482,320 |
|---|---|
| Length | 55 |
| Sequences | 9 |
| Columns | 56 |
| Reading direction | forward |
| Mean pairwise identity | 72.90 |
| Shannon entropy | 0.57021 |
| G+C content | 0.30165 |
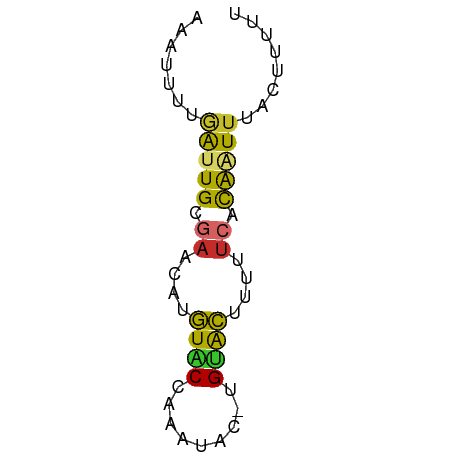
| Mean single sequence MFE | -12.01 |
| Consensus MFE | -6.53 |
| Energy contribution | -6.54 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.64 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.87 |
| SVM RNA-class probability | 0.999415 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
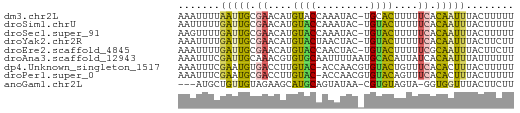
>dm3.chr2L 21482265 55 + 23011544 AAAAAGUAAAUUGUGAAAAAGUGCA-GUAUUUGGUACAUGUUCGCAAUUAAAAUUU ........(((((((((...((((.-.......))))...)))))))))....... ( -12.60, z-score = -3.36, R) >droSim1.chrU 3688789 55 - 15797150 AAAAAGUAAAUUGUGAAAAAGUACA-GUAUUUGGUACAUGUUCGCAAUCAAAAAUU .........((((((((...((((.-.......))))...))))))))........ ( -11.10, z-score = -3.39, R) >droSec1.super_91 63660 55 + 125313 AAAAAGUAAAUUGUGAAAAAGUACA-GUAUUUGGUACAUGUUCGCAAUCAAAACUU ...((((..((((((((...((((.-.......))))...))))))))....)))) ( -11.80, z-score = -3.12, R) >droYak2.chr2R 19830614 55 - 21139217 AAGAAGUAAAUUGUGAAAAAGUACA-GUAGUUAGUACAUGUUCGCAAUCAAAAUUU .........((((((((...((((.-.......))))...))))))))........ ( -11.20, z-score = -3.68, R) >droEre2.scaffold_4845 19550334 55 - 22589142 AAGAAGUAAAUUGCGAAAAAGUACA-GUAGUUGGUACAUGUUCGCAAUCAAAAUUU .........((((((((...((((.-.......))))...))))))))........ ( -13.00, z-score = -3.82, R) >droAna3.scaffold_12943 4888479 56 - 5039921 AAAAAAUAAAUUGUGAUAAUGUGCAUUAAAAUUGCACACGUUUGCAAUCGAAAUUU .........((((..(...((((((.......))))))...)..))))........ ( -12.30, z-score = -1.96, R) >dp4.Unknown_singleton_1517 5249 55 + 24252 AAAAAGUAAAGUGUGAAACAGUACACGUUGGU-GUACAAGGUCACAUUCGAAAUUU .........(((((((....((((((....))-))))....)))))))........ ( -14.80, z-score = -3.32, R) >droPer1.super_0 3264313 55 + 11822988 AAAAAGUAAAGUGUGAAACUGUACACGUUGGU-GUACAAGGUCGCAUUCGAAAUUU .........(((((((...(((((((....))-)))))...)))))))........ ( -16.10, z-score = -3.48, R) >anoGam1.chr2L 35943958 51 - 48795086 AAGAAGUAAACCACC-UACUACACG-UUAUACUGCAUGCUUCUACAACAGCAU--- .(((((((.......-.((.....)-).........)))))))..........--- ( -5.23, z-score = -0.48, R) >consensus AAAAAGUAAAUUGUGAAAAAGUACA_GUAGUUGGUACAUGUUCGCAAUCAAAAUUU .........(((((((....((((.........))))....)))))))........ ( -6.53 = -6.54 + 0.02)
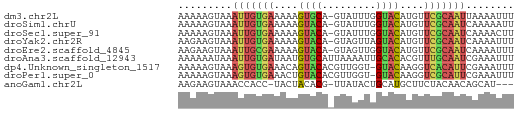
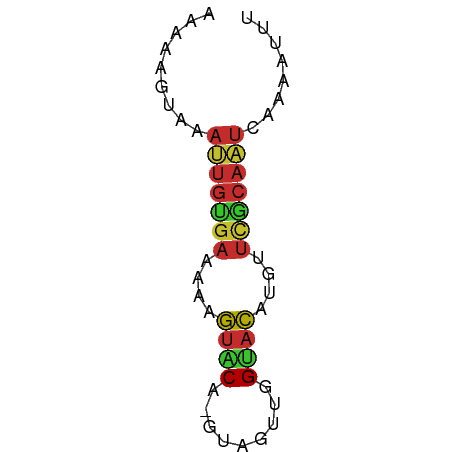
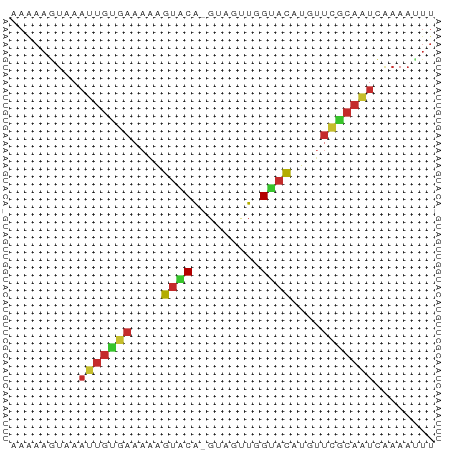
| Location | 21,482,265 – 21,482,320 |
|---|---|
| Length | 55 |
| Sequences | 9 |
| Columns | 56 |
| Reading direction | reverse |
| Mean pairwise identity | 72.90 |
| Shannon entropy | 0.57021 |
| G+C content | 0.30165 |
| Mean single sequence MFE | -9.54 |
| Consensus MFE | -3.09 |
| Energy contribution | -2.89 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.82 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.957902 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 21482265 55 - 23011544 AAAUUUUAAUUGCGAACAUGUACCAAAUAC-UGCACUUUUUCACAAUUUACUUUUU .......(((((.(((..((((........-))))....))).)))))........ ( -4.30, z-score = -1.44, R) >droSim1.chrU 3688789 55 + 15797150 AAUUUUUGAUUGCGAACAUGUACCAAAUAC-UGUACUUUUUCACAAUUUACUUUUU .......(((((.(((...((((.......-.))))...))).)))))........ ( -7.80, z-score = -3.05, R) >droSec1.super_91 63660 55 - 125313 AAGUUUUGAUUGCGAACAUGUACCAAAUAC-UGUACUUUUUCACAAUUUACUUUUU ((((...(((((.(((...((((.......-.))))...))).))))).))))... ( -10.10, z-score = -3.07, R) >droYak2.chr2R 19830614 55 + 21139217 AAAUUUUGAUUGCGAACAUGUACUAACUAC-UGUACUUUUUCACAAUUUACUUCUU .......(((((.(((...((((.......-.))))...))).)))))........ ( -9.00, z-score = -4.24, R) >droEre2.scaffold_4845 19550334 55 + 22589142 AAAUUUUGAUUGCGAACAUGUACCAACUAC-UGUACUUUUUCGCAAUUUACUUCUU .......(((((((((...((((.......-.))))...)))))))))........ ( -13.20, z-score = -5.85, R) >droAna3.scaffold_12943 4888479 56 + 5039921 AAAUUUCGAUUGCAAACGUGUGCAAUUUUAAUGCACAUUAUCACAAUUUAUUUUUU .......(((((.....(((((((.......))))))).....)))))........ ( -9.80, z-score = -1.65, R) >dp4.Unknown_singleton_1517 5249 55 - 24252 AAAUUUCGAAUGUGACCUUGUAC-ACCAACGUGUACUGUUUCACACUUUACUUUUU ..........(((((....((((-((....))))))....)))))........... ( -12.10, z-score = -2.91, R) >droPer1.super_0 3264313 55 - 11822988 AAAUUUCGAAUGCGACCUUGUAC-ACCAACGUGUACAGUUUCACACUUUACUUUUU ..........((.((..((((((-((....))))))))..)).))........... ( -11.00, z-score = -2.77, R) >anoGam1.chr2L 35943958 51 + 48795086 ---AUGCUGUUGUAGAAGCAUGCAGUAUAA-CGUGUAGUA-GGUGGUUUACUUCUU ---((((((((((....))).)))))))..-...((((..-......))))..... ( -8.60, z-score = 0.69, R) >consensus AAAUUUUGAUUGCGAACAUGUACCAAAUAC_UGUACUUUUUCACAAUUUACUUUUU .......(((((.((....((((.........))))....)).)))))........ ( -3.09 = -2.89 + -0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:56:27 2011