| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,477,255 – 21,477,366 |
| Length | 111 |
| Max. P | 0.996640 |

| Location | 21,477,255 – 21,477,366 |
|---|---|
| Length | 111 |
| Sequences | 13 |
| Columns | 122 |
| Reading direction | forward |
| Mean pairwise identity | 55.29 |
| Shannon entropy | 0.97959 |
| G+C content | 0.30463 |
| Mean single sequence MFE | -20.49 |
| Consensus MFE | -5.02 |
| Energy contribution | -5.49 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.96 |
| SVM RNA-class probability | 0.996640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
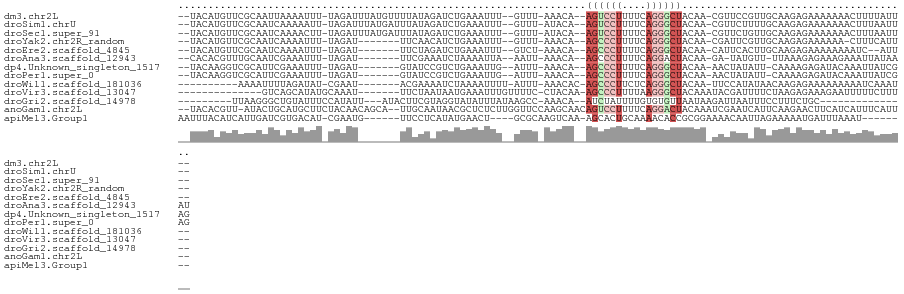
>dm3.chr2L 21477255 111 + 23011544 --UACAUGUUCGCAAUUAAAAUUU-UAGAUUUAUGUUUUAUAGAUCUGAAAUUU--GUUU-AAACA--AGUCCUUUUCAGGGCUACAA-CGUUCCGUUGCAAGAGAAAAAAACUUUUAUU-- --.........(((((..((((((-((((((((((...))))))))))))))))--....-.....--((((((....))))))....-......)))))(((((.......)))))...-- ( -24.40, z-score = -2.87, R) >droSim1.chrU 3629644 111 - 15797150 --UACAUGUUCGCAAUCAAAAAUU-UAGAUUUAUGAUUUAUAGAUCUGAAAUUU--GUUU-AUACA--AGUCCUUUUCAGGGCUACAA-CGUUCUUUUGCAAGAGAAAAAAACUUUAAUU-- --.........((((.((((..((-((((((((((...)))))))))))).)))--)...-.....--((((((....))))))....-.......))))....................-- ( -19.20, z-score = -1.27, R) >droSec1.super_91 77836 111 + 125313 --UACAUGUUCGCAAUCAAAACUU-UAGAUUUAUGAUUUAUAGAUCUGAAAUUU--GUUU-AUACA--AGUCCUUUUCAGGGCUACAA-CGUUCUGUUGCAAGAGAAAAAAACUUUAAUU-- --.........((((.((.(((((-((((((((((...))))))))))))....--....-.....--((((((....))))))....-.))).))))))....................-- ( -22.00, z-score = -1.89, R) >droYak2.chr2R_random 60139 103 - 67238 --UACAUGUUCGCAAUCAAAAUUU-UAGAU-------UUCAACAUCUGAAAUUU--GUUU-AAACA--AGCCCUUUUCAGGGCUACAA-CGAUUCGUUGCAAGAGAAAAAA-CUUUCAUU-- --....(((.((.((((.((((((-(((((-------......)))))))))))--....-.....--((((((....))))))....-.))))))..))).((((.....-.))))...-- ( -24.30, z-score = -3.66, R) >droEre2.scaffold_4845 19579535 102 - 22589142 --UACAUGUUCGCAAUCAAAAUUU-UAGAU-------UUCUAGAUCUGAAAUUU--GUCU-AAACA--AGCCCUUUUCAGGGCUACAA-CAUUCACUUGCAAGAGAAAAAAAAUC--AUU-- --.........((((...((((((-(((((-------(....))))))))))))--....-.....--((((((....))))))....-.......))))...............--...-- ( -22.60, z-score = -3.27, R) >droAna3.scaffold_12943 4888513 104 - 5039921 --CACACGUUUGCAAUCGAAAUUU-UAGAU-------UUCGAAAUCUAAAAUUA--AAUU-AAACA--AGCCCUUUUCAGGACUACAA-GA-UAUGUU-UUAAAGAGAAAGAAAUUAUAAAU --.....(((((.......(((((-(((((-------(....))))))))))).--....-...))--))).(((((.(((((.....-..-...)))-)).)))))............... ( -14.96, z-score = -1.49, R) >dp4.Unknown_singleton_1517 10412 105 + 24252 --UACAAGGUCGCAUUCGAAAUUU-UAGAU-------GUAUCCGUCUGAAAUUG--AUUU-AAACA--AGCCCUUUUCAGGGCUACAA-AACUAUAUU-CAAAAGAGAUACAAAUUAUCGAG --.............(((((((((-(((((-------(....))))))))))).--....-.....--((((((....))))))....-.........-..................)))). ( -22.20, z-score = -2.61, R) >droPer1.super_0 3286678 105 + 11822988 --UACAAGGUCGCAUUCGAAAUUU-UAGAU-------GUAUCCGUCUGAAAUUG--AUUU-AAACA--AGCCCUUUUCAGGGCUACAA-AACUAUAUU-CAAAAGAGAUACAAAUUAUCGAG --.............(((((((((-(((((-------(....))))))))))).--....-.....--((((((....))))))....-.........-..................)))). ( -22.20, z-score = -2.61, R) >droWil1.scaffold_181036 785 98 + 110856 ----------AAAAUUUUAGAUAU-CGAAU-------ACGAAAAUCUAAAAUUUU-AUUU-AAACAC-AGCCCUUCUCAGGGCUACAA-UUCCAUAUAACAAGAGAAAAAAAAAUCAAAU-- ----------((((((((((((.(-((...-------.)))..))))))))))))-....-......-((((((....))))))....-...............................-- ( -19.20, z-score = -5.29, R) >droVir3.scaffold_13047 1595517 97 + 19223366 --------------GUCAGCAUAUGCAAAU-------UUCUAAUAAUGAAAUUUGUUUUC-CUACAA-AGCCCUUUUAAGGGCUACAAAUACGAUUUUCUAAGAGAAAGAAUUUUUCUUU-- --------------((.((.....((((((-------(((.......)))))))))....-))))..-((((((....))))))................(((((((.....))))))).-- ( -20.50, z-score = -2.52, R) >droGri2.scaffold_14978 501459 92 + 1124632 ---------UUAAGGGCUGUAUUUCCAUAUU---AUACUUCGUAGGUAUAUUUAUAAGCC-AAACA--AUCUAUUUUGUGUGUUAAUAAGAUUAAUUUCCUUUCUGC--------------- ---------..(((((..............(---(((((.....))))))(((((.(((.-..(((--(......))))..))).))))).......))))).....--------------- ( -11.40, z-score = 0.56, R) >anoGam1.chr2L 35971879 115 - 48795086 --UACACGUU-AUACUGCAUGCUUCUACAACAGCA--UUGCAAUAACGCUCUCUUGGUUCCAAGCAACAGUCCUUUUCAGGACUACAAAUCGAAUCAUUCAAGAACUUCAUCAUUUCAUU-- --....((((-((..((((((((........))))--.)))))))))).(((..((((((........((((((....)))))).......))))))....)))................-- ( -23.96, z-score = -4.06, R) >apiMel3.Group1 12264803 102 - 25854376 AAUUUACAUCAUUGAUCGUGACAU-CGAAUG------UUCCUCAUAUGAACU----GCGCAAGUCAA-AGCACUGCAAAACACCGCGGAAAACAAUUAGAAAAAUGAUUUAAAU-------- .(((((.((((((..((.((((..-((...(------(((.......)))).----.))...)))).-....((((........))))..........))..)))))).)))))-------- ( -19.50, z-score = -1.39, R) >consensus __UACAUGUUCGCAAUCAAAAUUU_UAGAU_______UUAUAAAUCUGAAAUUU__GUUU_AAACA__AGCCCUUUUCAGGGCUACAA_CAUUAUGUUGCAAGAGAAAAAAAAUUUAAUU__ ....................................................................((((((....))))))...................................... ( -5.02 = -5.49 + 0.47)

| Location | 21,477,255 – 21,477,366 |
|---|---|
| Length | 111 |
| Sequences | 13 |
| Columns | 122 |
| Reading direction | reverse |
| Mean pairwise identity | 55.29 |
| Shannon entropy | 0.97959 |
| G+C content | 0.30463 |
| Mean single sequence MFE | -21.80 |
| Consensus MFE | -6.59 |
| Energy contribution | -6.68 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.83 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.989602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
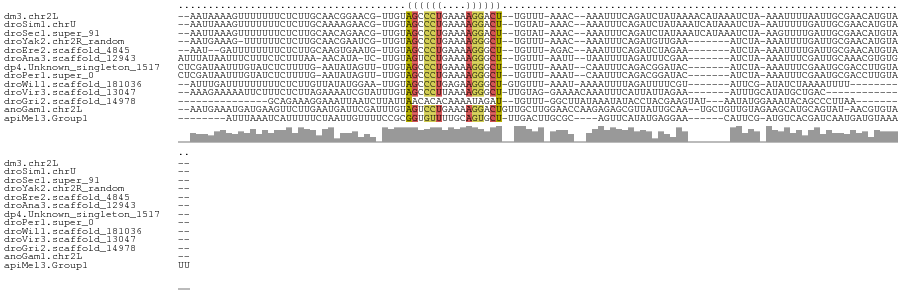
>dm3.chr2L 21477255 111 - 23011544 --AAUAAAAGUUUUUUUCUCUUGCAACGGAACG-UUGUAGCCCUGAAAAGGACU--UGUUU-AAAC--AAAUUUCAGAUCUAUAAAACAUAAAUCUA-AAAUUUUAAUUGCGAACAUGUA-- --.....(((((((((((..((((((((...))-))))))....))))))))))--)((((-.((.--((((((.((((.(((.....))).)))).-))))))...))..)))).....-- ( -21.00, z-score = -2.13, R) >droSim1.chrU 3629644 111 + 15797150 --AAUUAAAGUUUUUUUCUCUUGCAAAAGAACG-UUGUAGCCCUGAAAAGGACU--UGUAU-AAAC--AAAUUUCAGAUCUAUAAAUCAUAAAUCUA-AAUUUUUGAUUGCGAACAUGUA-- --.....(((((((((((..((((((.......-))))))....))))))))))--)(((.-....--((((((.((((.(((.....))).)))))-))))).....))).........-- ( -16.90, z-score = -0.59, R) >droSec1.super_91 77836 111 - 125313 --AAUUAAAGUUUUUUUCUCUUGCAACAGAACG-UUGUAGCCCUGAAAAGGACU--UGUAU-AAAC--AAAUUUCAGAUCUAUAAAUCAUAAAUCUA-AAGUUUUGAUUGCGAACAUGUA-- --.....(((((((((((..(((((((.....)-))))))....))))))))))--)(((.-....--((((((.((((.(((.....))).)))).-))))))....))).........-- ( -21.00, z-score = -1.63, R) >droYak2.chr2R_random 60139 103 + 67238 --AAUGAAAG-UUUUUUCUCUUGCAACGAAUCG-UUGUAGCCCUGAAAAGGGCU--UGUUU-AAAC--AAAUUUCAGAUGUUGAA-------AUCUA-AAAUUUUGAUUGCGAACAUGUA-- --..(((((.-..........(((((((...))-)))))(((((....))))).--.....-....--...))))).(((((..(-------(((..-.......))))...)))))...-- ( -23.90, z-score = -2.07, R) >droEre2.scaffold_4845 19579535 102 + 22589142 --AAU--GAUUUUUUUUCUCUUGCAAGUGAAUG-UUGUAGCCCUGAAAAGGGCU--UGUUU-AGAC--AAAUUUCAGAUCUAGAA-------AUCUA-AAAUUUUGAUUGCGAACAUGUA-- --...--.............((((((.....((-((..((((((....))))))--.....-.)))--)....(((((..(((..-------..)))-....))))))))))).......-- ( -22.20, z-score = -1.35, R) >droAna3.scaffold_12943 4888513 104 + 5039921 AUUUAUAAUUUCUUUCUCUUUAA-AACAUA-UC-UUGUAGUCCUGAAAAGGGCU--UGUUU-AAUU--UAAUUUUAGAUUUCGAA-------AUCUA-AAAUUUCGAUUGCAAACGUGUG-- .......................-..((((-(.-((((((((((....))))))--.....-((((--.(((((((((((....)-------)))))-)))))..))))))))..)))))-- ( -20.30, z-score = -2.93, R) >dp4.Unknown_singleton_1517 10412 105 - 24252 CUCGAUAAUUUGUAUCUCUUUUG-AAUAUAGUU-UUGUAGCCCUGAAAAGGGCU--UGUUU-AAAU--CAAUUUCAGACGGAUAC-------AUCUA-AAAUUUCGAAUGCGACCUUGUA-- .((((.....(((((((..((((-((....(((-(...((((((....))))))--.....-))))--....)))))).))))))-------)....-.....)))).............-- ( -25.94, z-score = -2.93, R) >droPer1.super_0 3286678 105 - 11822988 CUCGAUAAUUUGUAUCUCUUUUG-AAUAUAGUU-UUGUAGCCCUGAAAAGGGCU--UGUUU-AAAU--CAAUUUCAGACGGAUAC-------AUCUA-AAAUUUCGAAUGCGACCUUGUA-- .((((.....(((((((..((((-((....(((-(...((((((....))))))--.....-))))--....)))))).))))))-------)....-.....)))).............-- ( -25.94, z-score = -2.93, R) >droWil1.scaffold_181036 785 98 - 110856 --AUUUGAUUUUUUUUUCUCUUGUUAUAUGGAA-UUGUAGCCCUGAGAAGGGCU-GUGUUU-AAAU-AAAAUUUUAGAUUUUCGU-------AUUCG-AUAUCUAAAAUUUU---------- --((((((..........(((........))).-...(((((((....))))))-)...))-))))-((((((((((((..(((.-------...))-).))))))))))))---------- ( -23.60, z-score = -3.54, R) >droVir3.scaffold_13047 1595517 97 - 19223366 --AAAGAAAAAUUCUUUCUCUUAGAAAAUCGUAUUUGUAGCCCUUAAAAGGGCU-UUGUAG-GAAAACAAAUUUCAUUAUUAGAA-------AUUUGCAUAUGCUGAC-------------- --..(((((.....))))).((((....((.(((....((((((....))))))-..))).-))...((((((((.......)))-------)))))......)))).-------------- ( -20.60, z-score = -2.02, R) >droGri2.scaffold_14978 501459 92 - 1124632 ---------------GCAGAAAGGAAAUUAAUCUUAUUAACACACAAAAUAGAU--UGUUU-GGCUUAUAAAUAUACCUACGAAGUAU---AAUAUGGAAAUACAGCCCUUAA--------- ---------------.....((((................((.((((......)--))).)-)(((......(((((.......))))---).(((....))).)))))))..--------- ( -9.10, z-score = 0.52, R) >anoGam1.chr2L 35971879 115 + 48795086 --AAUGAAAUGAUGAAGUUCUUGAAUGAUUCGAUUUGUAGUCCUGAAAAGGACUGUUGCUUGGAACCAAGAGAGCGUUAUUGCAA--UGCUGUUGUAGAAGCAUGCAGUAU-AACGUGUA-- --..((.(((((((...((((((...(.(((((.(..(((((((....)))))))..).))))).)))))))..))))))).))(--(((((((((....))).)))))))-........-- ( -32.70, z-score = -3.22, R) >apiMel3.Group1 12264803 102 + 25854376 --------AUUUAAAUCAUUUUUCUAAUUGUUUUCCGCGGUGUUUUGCAGUGCU-UUGACUUGCGC----AGUUCAUAUGAGGAA------CAUUCG-AUGUCACGAUCAAUGAUGUAAAUU --------(((((.((((((..((..((((..((((((((....)))).((((.-.......))))----...........))))------....))-)).....))..)))))).))))). ( -20.20, z-score = 0.10, R) >consensus __AAUUAAAUUUUUUUUCUCUUGCAAAAUAAUG_UUGUAGCCCUGAAAAGGGCU__UGUUU_AAAC__AAAUUUCAGAUCUAGAA_______AUCUA_AAAUUUUGAUUGCGAACAUGUA__ ......................................((((((....)))))).................................................................... ( -6.59 = -6.68 + 0.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:56:26 2011