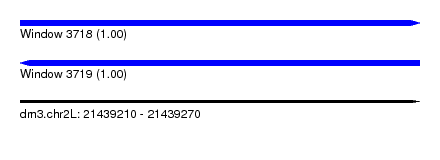
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,439,210 – 21,439,270 |
| Length | 60 |
| Max. P | 0.998915 |

| Location | 21,439,210 – 21,439,270 |
|---|---|
| Length | 60 |
| Sequences | 8 |
| Columns | 67 |
| Reading direction | forward |
| Mean pairwise identity | 68.50 |
| Shannon entropy | 0.65104 |
| G+C content | 0.32766 |
| Mean single sequence MFE | -9.81 |
| Consensus MFE | -8.40 |
| Energy contribution | -8.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.997273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
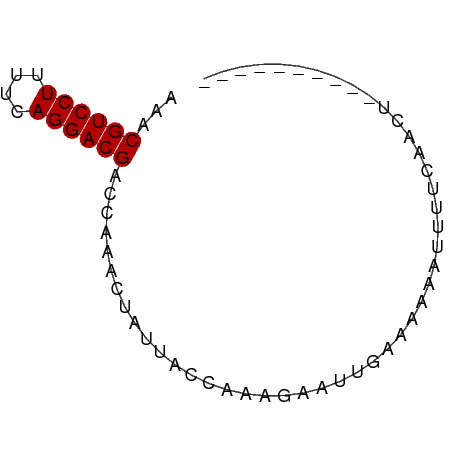
>dm3.chr2L 21439210 60 + 23011544 AACCGUCCUUUUCAGGACGACCAAAUUAUUACCAAAGGAUUGAAAAAUUUUUUAGCUUGG------- ...((((((....)))))).((((.(((......(((((((....)))))))))).))))------- ( -12.50, z-score = -1.94, R) >droGri2.scaffold_14978 506109 55 + 1124632 AAACGUCCUUUUCAGGACGACCAAAAUAUGU--AAAAGA--GAUUUAAUUUUUCAUUGA-------- ...((((((....))))))............--....((--((.......)))).....-------- ( -9.90, z-score = -1.61, R) >droWil1.scaffold_184887 3 65 - 1286 AACCGUCCUUUUCAGGACGACCAAAAAUUAA--AAGAAAUUAAUACAGUUUUCCAUCCAUGCAUAUA ...((((((....))))))............--.................................. ( -8.30, z-score = -1.97, R) >droPer1.super_0 3281364 54 + 11822988 AAACGUCCUUUUCAGGACGACCAAACUAUGACAAAAGAAUUAAGAGAAUUUUCA------------- ...((((((....))))))...................................------------- ( -8.50, z-score = -1.31, R) >dp4.Unknown_singleton_1517 23568 54 + 24252 AAACGUCCUUUUCAGGACGACCAAACUAUGACUAAAGAAUUAAGAGAAUUUUCA------------- ...((((((....))))))...................................------------- ( -8.50, z-score = -1.21, R) >droAna3.scaffold_8577 7840 64 - 7985 AAACGUCCUUUUCAGGACGACCACACUUUUUCAAAAGAAUAUA---AAUUUUCAACUACAAGAUUAG ...((((((....)))))).................(((....---....))).............. ( -9.20, z-score = -2.68, R) >droYak2.chrU 9058519 64 - 28119190 AACCGUCCUUUUCAGGACGACCAAAUUGUUACCAAAGAUUUGAAAAAUUUUUUAACUACGUUAC--- ...((((((....))))))........((((..(((((((.....)))))))))))........--- ( -10.50, z-score = -1.49, R) >droSec1.super_91 118176 60 + 125313 AACCGUCCUUUUCAGGACGACCAAAUUAUUACCAAAGAAUUGGAAAAAUUUUACACUUGG------- ...((((((....))))))............((((((((((.....))))))....))))------- ( -11.10, z-score = -1.65, R) >consensus AAACGUCCUUUUCAGGACGACCAAACUAUUACCAAAGAAUUGAAAAAAUUUUCAACU__________ ...((((((....))))))................................................ ( -8.40 = -8.40 + 0.00)

| Location | 21,439,210 – 21,439,270 |
|---|---|
| Length | 60 |
| Sequences | 8 |
| Columns | 67 |
| Reading direction | reverse |
| Mean pairwise identity | 68.50 |
| Shannon entropy | 0.65104 |
| G+C content | 0.32766 |
| Mean single sequence MFE | -12.90 |
| Consensus MFE | -10.75 |
| Energy contribution | -10.75 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.55 |
| SVM RNA-class probability | 0.998915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
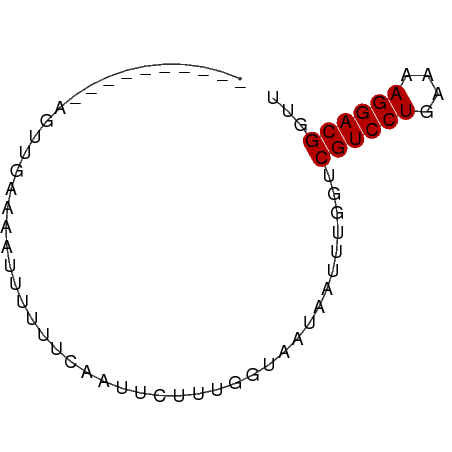
>dm3.chr2L 21439210 60 - 23011544 -------CCAAGCUAAAAAAUUUUUCAAUCCUUUGGUAAUAAUUUGGUCGUCCUGAAAAGGACGGUU -------....((((((..(((..((((....)))).)))..))))))((((((....))))))... ( -14.90, z-score = -2.49, R) >droGri2.scaffold_14978 506109 55 - 1124632 --------UCAAUGAAAAAUUAAAUC--UCUUUU--ACAUAUUUUGGUCGUCCUGAAAAGGACGUUU --------..................--......--............((((((....))))))... ( -10.40, z-score = -1.83, R) >droWil1.scaffold_184887 3 65 + 1286 UAUAUGCAUGGAUGGAAAACUGUAUUAAUUUCUU--UUAAUUUUUGGUCGUCCUGAAAAGGACGGUU ..........(((.((((.....(((((......--))))))))).)))(((((....))))).... ( -13.20, z-score = -1.56, R) >droPer1.super_0 3281364 54 - 11822988 -------------UGAAAAUUCUCUUAAUUCUUUUGUCAUAGUUUGGUCGUCCUGAAAAGGACGUUU -------------...................................((((((....))))))... ( -10.40, z-score = -1.53, R) >dp4.Unknown_singleton_1517 23568 54 - 24252 -------------UGAAAAUUCUCUUAAUUCUUUAGUCAUAGUUUGGUCGUCCUGAAAAGGACGUUU -------------...................................((((((....))))))... ( -10.40, z-score = -1.47, R) >droAna3.scaffold_8577 7840 64 + 7985 CUAAUCUUGUAGUUGAAAAUU---UAUAUUCUUUUGAAAAAGUGUGGUCGUCCUGAAAAGGACGUUU (((..(((.(((..(((....---....)))..)))...)))..))).((((((....))))))... ( -13.00, z-score = -2.50, R) >droYak2.chrU 9058519 64 + 28119190 ---GUAACGUAGUUAAAAAAUUUUUCAAAUCUUUGGUAACAAUUUGGUCGUCCUGAAAAGGACGGUU ---.....................((((((...((....))))))))(((((((....))))))).. ( -13.00, z-score = -1.81, R) >droSec1.super_91 118176 60 - 125313 -------CCAAGUGUAAAAUUUUUCCAAUUCUUUGGUAAUAAUUUGGUCGUCCUGAAAAGGACGGUU -------((((((.....(((...((((....)))).))).))))))(((((((....))))))).. ( -17.90, z-score = -3.78, R) >consensus __________AGUUGAAAAUUUUUUCAAUUCUUUGGUAAUAAUUUGGUCGUCCUGAAAAGGACGGUU ................................................((((((....))))))... (-10.75 = -10.75 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:56:22 2011