| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,423,135 – 21,423,190 |
| Length | 55 |
| Max. P | 0.999859 |

| Location | 21,423,135 – 21,423,190 |
|---|---|
| Length | 55 |
| Sequences | 11 |
| Columns | 66 |
| Reading direction | forward |
| Mean pairwise identity | 58.05 |
| Shannon entropy | 0.88502 |
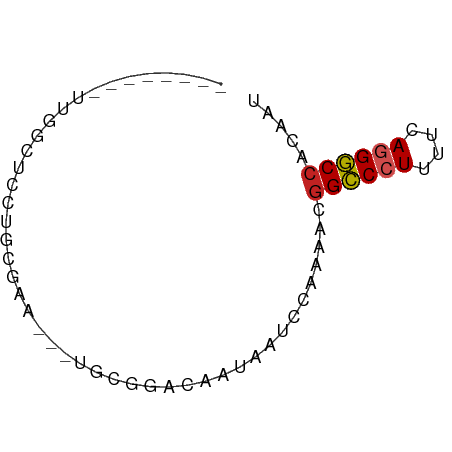
| G+C content | 0.46956 |
| Mean single sequence MFE | -18.47 |
| Consensus MFE | -11.74 |
| Energy contribution | -13.01 |
| Covariance contribution | 1.27 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.60 |
| SVM RNA-class probability | 0.999859 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
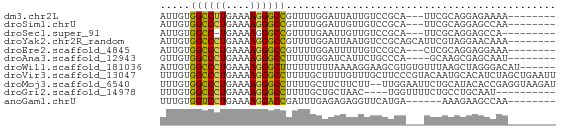
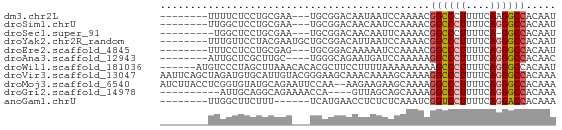
>dm3.chr2L 21423135 55 + 23011544 AUUGUGGCCUUGAAAAGGGCCGUUUUGGAUUAUUGUCCGCA---UUCGCAGGAGAAAA-------- .((((((((((....)))))).....((((....))))...---...)))).......-------- ( -17.40, z-score = -1.81, R) >droSim1.chrU 65411 55 - 15797150 AUUGUGGCCCUGAAAAGGGCCGUUUUGGAUUGUUGUCCGCA---UUCGCAGGAGCCAA-------- .....((((((....))))))(((((((((....))))((.---...)).)))))...-------- ( -21.60, z-score = -2.34, R) >droSec1.super_91 64049 53 + 125313 AUUGUGGCC-UGAAAAGGGCCGUUUUGAAUUGUUGUCCGCA---UUCGCAGGAGCCA--------- .....((((-(.....)))))..............(((((.---...)).)))....--------- ( -13.10, z-score = 0.18, R) >droYak2.chr2R_random 60460 58 - 67238 AUUGUGGCCCUGAAAAGGGCCGUUUUGGAUUAAUGUCCGCAGCAUUCGUAGGAACAAA-------- .((((((((((....))))))(((.(((((....))))).)))..........)))).-------- ( -23.20, z-score = -3.62, R) >droEre2.scaffold_4845 19535491 55 - 22589142 AUUGUGGCCCUGAAAAGGGCCGUUUUGGAUUUUUGUCCGCA---CUCGCAGGAGGAAA-------- ..(((((((((....)))))).....((((....)))))))---(((....)))....-------- ( -21.40, z-score = -2.38, R) >droAna3.scaffold_12943 4809564 54 - 5039921 GUUGUGGCCCUGAAAAGGGCCUUUUUGGAUCAUUCUGCCCA----GCAAGCGAGCAAU-------- ((((.((((((....)))))).....(((....)))...))----))...........-------- ( -17.50, z-score = -0.99, R) >droWil1.scaffold_181036 1021 60 + 110856 AUUGUGGCCCUGAAAAGGGCUUUUUUUUUAAAAGGAAGCGUGUGUUUAAGCUAGGGACAU------ .((.(((((((....)))((((((((.....))))))))..........)))).))....------ ( -14.80, z-score = -0.85, R) >droVir3.scaffold_13047 1585291 66 + 19223366 UUUGUGGCCCUGAAAAGGGCCUUUUGCUUUUGUUUGCUUCCCGUACAAUGCACAUCUAGCUGAAUU .....((((((....))))))(((.(((...((.(((............))).))..))).))).. ( -18.20, z-score = -1.84, R) >droMoj3.scaffold_6540 26857080 64 - 34148556 UUUGUGGCCCUGAAAAGGGCCUUUUGCUUCUUCUU--UUGGAAUUCUGCAUACACCGAGGUAAGAU .....((((((....))))))(((((((((.....--.((.(....).))......))))))))). ( -20.80, z-score = -2.43, R) >droGri2.scaffold_14978 482894 52 + 1124632 UUUGUGGCCCUGAAAAGGGCCUUUUGCUGCUAAC----UGGUUUUCUGCCUGCAAU---------- .....((((((....))))))......(((....----.(((.....))).)))..---------- ( -18.20, z-score = -2.45, R) >anoGam1.chrU 44504713 52 + 59568033 UUUGUGGUCCUGAAAAGGACCGAUUUGAGAGAGGUUCAUGA------AAAGAAGCCAA-------- ....(((((((....)))))))..........((((..(..------..)..))))..-------- ( -17.00, z-score = -3.59, R) >consensus AUUGUGGCCCUGAAAAGGGCCGUUUUGGAUUAUUGUCCGCA___UUCGCAGGAGCCAA________ .....((((((....))))))............................................. (-11.74 = -13.01 + 1.27)
| Location | 21,423,135 – 21,423,190 |
|---|---|
| Length | 55 |
| Sequences | 11 |
| Columns | 66 |
| Reading direction | reverse |
| Mean pairwise identity | 58.05 |
| Shannon entropy | 0.88502 |
| G+C content | 0.46956 |
| Mean single sequence MFE | -14.25 |
| Consensus MFE | -10.06 |
| Energy contribution | -10.16 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999519 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
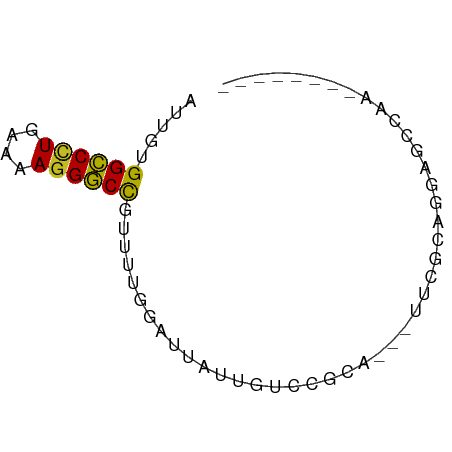
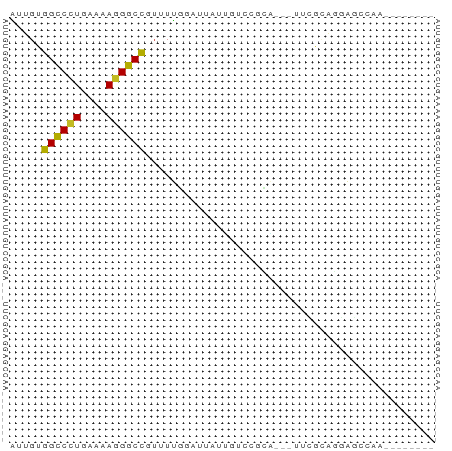
>dm3.chr2L 21423135 55 - 23011544 --------UUUUCUCCUGCGAA---UGCGGACAAUAAUCCAAAACGGCCCUUUUCAAGGCCACAAU --------.......((((...---.))))...............((((........))))..... ( -11.20, z-score = -1.25, R) >droSim1.chrU 65411 55 + 15797150 --------UUGGCUCCUGCGAA---UGCGGACAACAAUCCAAAACGGCCCUUUUCAGGGCCACAAU --------(((..(((.((...---.)))))...)))........((((((....))))))..... ( -16.30, z-score = -1.62, R) >droSec1.super_91 64049 53 - 125313 ---------UGGCUCCUGCGAA---UGCGGACAACAAUUCAAAACGGCCCUUUUCA-GGCCACAAU ---------......((((...---.))))...............((((.......-))))..... ( -10.90, z-score = -0.27, R) >droYak2.chr2R_random 60460 58 + 67238 --------UUUGUUCCUACGAAUGCUGCGGACAUUAAUCCAAAACGGCCCUUUUCAGGGCCACAAU --------(((((....)))))......(((......))).....((((((....))))))..... ( -15.40, z-score = -2.01, R) >droEre2.scaffold_4845 19535491 55 + 22589142 --------UUUCCUCCUGCGAG---UGCGGACAAAAAUCCAAAACGGCCCUUUUCAGGGCCACAAU --------.......((((...---.))))...............((((((....))))))..... ( -15.70, z-score = -1.81, R) >droAna3.scaffold_12943 4809564 54 + 5039921 --------AUUGCUCGCUUGC----UGGGCAGAAUGAUCCAAAAAGGCCCUUUUCAGGGCCACAAC --------.(((((((.....----))))))).............((((((....))))))..... ( -18.10, z-score = -1.59, R) >droWil1.scaffold_181036 1021 60 - 110856 ------AUGUCCCUAGCUUAAACACACGCUUCCUUUUAAAAAAAAAGCCCUUUUCAGGGCCACAAU ------.(((....(((..........)))................(((((....))))).))).. ( -8.00, z-score = -0.71, R) >droVir3.scaffold_13047 1585291 66 - 19223366 AAUUCAGCUAGAUGUGCAUUGUACGGGAAGCAAACAAAAGCAAAAGGCCCUUUUCAGGGCCACAAA ......(((...(((((...)))))...)))..............((((((....))))))..... ( -18.30, z-score = -1.74, R) >droMoj3.scaffold_6540 26857080 64 + 34148556 AUCUUACCUCGGUGUAUGCAGAAUUCCAA--AAGAAGAAGCAAAAGGCCCUUUUCAGGGCCACAAA ....(((....)))..(((....(((...--..)))...)))...((((((....))))))..... ( -15.10, z-score = -1.13, R) >droGri2.scaffold_14978 482894 52 - 1124632 ----------AUUGCAGGCAGAAAACCA----GUUAGCAGCAAAAGGCCCUUUUCAGGGCCACAAA ----------.((((..((.........----))..)))).....((((((....))))))..... ( -15.60, z-score = -1.78, R) >anoGam1.chrU 44504713 52 - 59568033 --------UUGGCUUCUUU------UCAUGAACCUCUCUCAAAUCGGUCCUUUUCAGGACCACAAA --------..((.(((...------....)))))...........((((((....))))))..... ( -12.10, z-score = -2.56, R) >consensus ________UUGGCUCCUGCGAA___UGCGGACAAUAAUCCAAAACGGCCCUUUUCAGGGCCACAAU .............................................((((((....))))))..... (-10.06 = -10.16 + 0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:56:20 2011