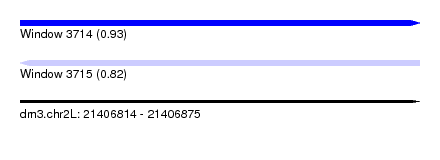
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,406,814 – 21,406,875 |
| Length | 61 |
| Max. P | 0.925031 |

| Location | 21,406,814 – 21,406,875 |
|---|---|
| Length | 61 |
| Sequences | 5 |
| Columns | 62 |
| Reading direction | forward |
| Mean pairwise identity | 62.07 |
| Shannon entropy | 0.67399 |
| G+C content | 0.45979 |
| Mean single sequence MFE | -9.66 |
| Consensus MFE | -6.24 |
| Energy contribution | -5.48 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.80 |
| Mean z-score | -0.56 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.925031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
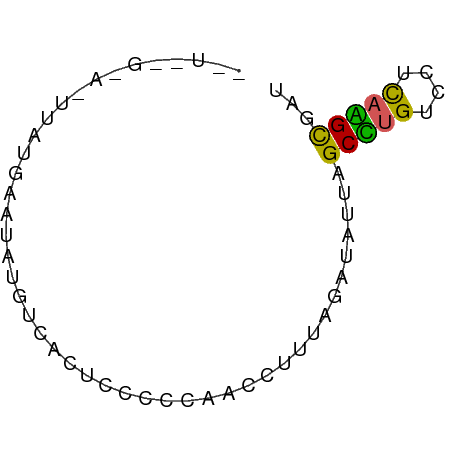
>dm3.chr2L 21406814 61 + 23011544 CUUAGGGACUUAUGGAUAUGUCACUCCCCCAACCUUU-GAAUUUGGCCUGUCCUCAGGUCGU ......(((((..(((((.((((..............-.....)))).)))))..))))).. ( -13.91, z-score = 0.10, R) >droYak2.chrU 21712311 60 + 28119190 CGUAUGAAGAAAUGAAUAAGUCACUCCCCCACCUUUUAGAUAU-AGCUUGUCCUCAAGCGA- ...........................................-.(((((....)))))..- ( -6.30, z-score = -0.16, R) >droPer1.super_57 135472 61 - 429542 UGUGGGGAAUUAUGGAUAUGUCACUCCCCCAACGUUU-GUUAUUAGCCUGUCCCUAGGCGAU ((.(((((....(((.....))).)))))))......-.......(((((....)))))... ( -14.20, z-score = 0.04, R) >droGri2.scaffold_6403 3511 53 + 10120 ---------AUCUGAAUAUGUCACUUCCCCAUCCAUUAGAAAUUGGCUUGUCCUCAAGCGUU ---------.(((((..(((.........)))...))))).....(((((....)))))... ( -7.60, z-score = -0.91, R) >droVir3.scaffold_12799 335169 52 - 516347 ---------UUGUGAAUAUGUCACUCCUCCCCACUGAAAAUAA-AGCGCGUCCUCCCGCCUU ---------..((((.....))))..................(-((.(((......)))))) ( -6.30, z-score = -1.87, R) >consensus __U__G_A_UUAUGAAUAUGUCACUCCCCCAACCUUUAGAUAUUAGCCUGUCCUCAAGCGAU .............................................(((((....)))))... ( -6.24 = -5.48 + -0.76)

| Location | 21,406,814 – 21,406,875 |
|---|---|
| Length | 61 |
| Sequences | 5 |
| Columns | 62 |
| Reading direction | reverse |
| Mean pairwise identity | 62.07 |
| Shannon entropy | 0.67399 |
| G+C content | 0.45979 |
| Mean single sequence MFE | -13.60 |
| Consensus MFE | -5.80 |
| Energy contribution | -5.36 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.823562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 21406814 61 - 23011544 ACGACCUGAGGACAGGCCAAAUUC-AAAGGUUGGGGGAGUGACAUAUCCAUAAGUCCCUAAG .((((((..(((.........)))-..))))))(((((.(............).)))))... ( -14.40, z-score = -0.01, R) >droYak2.chrU 21712311 60 - 28119190 -UCGCUUGAGGACAAGCU-AUAUCUAAAAGGUGGGGGAGUGACUUAUUCAUUUCUUCAUACG -..(((((....))))).-...........((((((((((((.....))))))))))))... ( -17.60, z-score = -3.19, R) >droPer1.super_57 135472 61 + 429542 AUCGCCUAGGGACAGGCUAAUAAC-AAACGUUGGGGGAGUGACAUAUCCAUAAUUCCCCACA ...((((......))))...((((-....))))(((((((............)))))))... ( -16.50, z-score = -1.17, R) >droGri2.scaffold_6403 3511 53 - 10120 AACGCUUGAGGACAAGCCAAUUUCUAAUGGAUGGGGAAGUGACAUAUUCAGAU--------- ...(((((....)))))..........((((((.(.......).))))))...--------- ( -8.10, z-score = 0.01, R) >droVir3.scaffold_12799 335169 52 + 516347 AAGGCGGGAGGACGCGCU-UUAUUUUCAGUGGGGAGGAGUGACAUAUUCACAA--------- ((((((.(....).))))-)).(((((......)))))((((.....))))..--------- ( -11.40, z-score = -1.59, R) >consensus AACGCCUGAGGACAAGCUAAUAUCUAAAGGGUGGGGGAGUGACAUAUUCAUAA_U_C__A__ ...(((((....)))))..................(((........)))............. ( -5.80 = -5.36 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:56:19 2011