
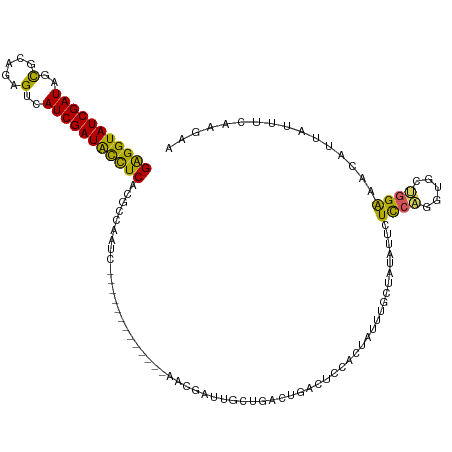
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,344,490 – 21,344,675 |
| Length | 185 |
| Max. P | 0.739373 |

| Location | 21,344,490 – 21,344,597 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.95 |
| Shannon entropy | 0.45785 |
| G+C content | 0.44064 |
| Mean single sequence MFE | -28.54 |
| Consensus MFE | -17.56 |
| Energy contribution | -16.12 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.605947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 21344490 107 - 23011544 GAGGUAUCGAUAGCGCAUGGUCAUCGAUACCUCACGCCAAUC-------------AACGAUUGCUGAAUGACUCCACUAUUUGUUAUCUUAUCCCGGUGCUGGAAACAUUAUUUCAAGAA (((((((((((..(.....)..))))))))))).((((..((-------------(........)))(((((..........)))))........)))).((....))............ ( -28.00, z-score = -1.86, R) >droEre2.scaffold_4845 19436754 120 + 22589142 GGGGUAUCGAUAGCGCAGGGUCAUCGAUGUUUCACGCCAAUCACGACAAUGCGAGAAUGCUCGCUAACUGUUUGCACUAUUUGCCAUUUUCUCCUCUUGCGGGGAAGAUUAUUUCAAGAU ..((((..(((((.((((((((...(((...........)))..)))...(((((....)))))......))))).)))))))))(((((((((......)))))))))........... ( -32.00, z-score = -0.46, R) >droYak2.chr2R 19709059 114 + 21139217 GGGGUAUCGAUAGCGAAGUGUCAUCGAUGUUUCACGCCAAUUACGACUAUGUGGGAAUGCUCGCAGACUGUUUCCACUAUGUGCUAU---CUUCUUGUGCGUGAAAGAAUAUUAGGC--- ((..(((((((..(.....)..)))))))..))..(((............((((((((..((...))..))))))))((((..(...---......)..))))...........)))--- ( -28.90, z-score = 0.19, R) >droSec1.super_18 1273259 107 - 1342155 GAGGUAUCGAUAGUGCAGAGUCAUCGAUACCUCACGCCAAUC-------------AUCGAUUGCUGACUGACUCCACUACUUCCUAUAUUUUCCAGGUGCUGGAAACAUUAUUUCAAGAA (((((((((((..(.....)..))))))))))).((.(((((-------------...))))).)).............(((...(((.(((((((...)))))))...)))...))).. ( -26.90, z-score = -1.72, R) >droSim1.chr2L 20945503 107 - 22036055 GAGGUAUCGAUAGUGCAGAGUCAUCGAUACCUCACGCCAAUC-------------AUCGAUUGUUGACUGACUCCACUACUUCCUAUAUUUUCCAGGUGCUGGAAACAUUAUUUCAAGAA (((((((((((..(.....)..))))))))))).((.(((((-------------...))))).)).............(((...(((.(((((((...)))))))...)))...))).. ( -26.90, z-score = -1.73, R) >consensus GAGGUAUCGAUAGCGCAGAGUCAUCGAUACCUCACGCCAAUC_____________AACGAUUGCUGACUGACUCCACUAUUUGCUAUAUUCUCCAGGUGCUGGAAACAUUAUUUCAAGAA (((((((((((..(.....)..)))))))))))..........................................................((((.....))))................ (-17.56 = -16.12 + -1.44)


| Location | 21,344,530 – 21,344,637 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.57 |
| Shannon entropy | 0.43587 |
| G+C content | 0.47226 |
| Mean single sequence MFE | -32.01 |
| Consensus MFE | -17.96 |
| Energy contribution | -20.52 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.739373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 21344530 107 + 23011544 AUAGUGGAGUCAUUCAGCAAUCG-------------UUGAUUGGCGUGAGGUAUCGAUGACCAUGCGCUAUCGAUACCUCCGUUGGAAUUAUCACAUCGUAGCUCUACUGUUUGUCAAGU ((((((((((...(((((....)-------------))))(..(((.((((((((((((.(.....).)))))))))))))))..)...............))))))))))......... ( -39.30, z-score = -3.72, R) >droEre2.scaffold_4845 19436794 115 - 22589142 AUAGUGCAAACAGUUAGCGAGCAUUCUCGCAUUGUCGUGAUUGGCGUGAAACAUCGAUGACCCUGCGCUAUCGAUACCCCUAUCGGA-CUAUCACAUCGCAGCUCUAAUGUAAAGU---- .(((.((..(((((..(((((....)))))))))).(((((((((((.................))))))..((((..((....)).-.))))..))))).)).))).........---- ( -27.53, z-score = 0.08, R) >droYak2.chr2R 19709093 116 - 21139217 AUAGUGGAAACAGUCUGCGAGCAUUCCCACAUAGUCGUAAUUGGCGUGAAACAUCGAUGACACUUCGCUAUCGAUACCCCCAUCGGAACUAUCACAUCUCAGCUCUACUGCGCUGU---- ((((((....((((....((((...........((((....))))((((...(((((((.(.....).))))))).((......)).....))))......)))).))))))))))---- ( -25.50, z-score = 0.14, R) >droSec1.super_18 1273299 101 + 1342155 GUAGUGGAGUCAGUCAGCAAUCG-------------AUGAUUGGCGUGAGGUAUCGAUGACUCUGCACUAUCGAUACCUCCCUUGGAACUAUCGCAUCGCAGCUCUACU-UAAGU----- ..((((((((((((((.......-------------.))))))(((.((((((((((((.........))))))))))))...((((....)).)).))).))))))))-.....----- ( -34.20, z-score = -2.15, R) >droSim1.chr2L 20945543 101 + 22036055 GUAGUGGAGUCAGUCAACAAUCG-------------AUGAUUGGCGUGAGGUAUCGAUGACUCUGCACUAUCGAUACCUCCCUUGGAACUAUCGCAUCGCAGCUCUACU-UAAGU----- ..((((((((((((((.(....)-------------.))))))(((.((((((((((((.........))))))))))))...((((....)).)).))).))))))))-.....----- ( -33.50, z-score = -2.41, R) >consensus AUAGUGGAGUCAGUCAGCAAUCG_____________AUGAUUGGCGUGAGGUAUCGAUGACCCUGCGCUAUCGAUACCUCCAUUGGAACUAUCACAUCGCAGCUCUACUGUAAGU_____ ((((((((((.......((((((..............))))))..((((((((((((((.........)))))))))).((...)).....))))......))))))))))......... (-17.96 = -20.52 + 2.56)


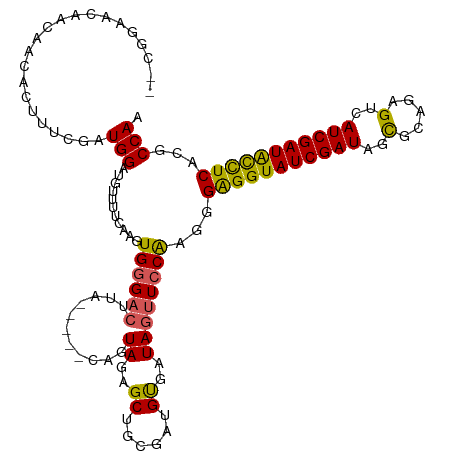
| Location | 21,344,557 – 21,344,675 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.13 |
| Shannon entropy | 0.29225 |
| G+C content | 0.46408 |
| Mean single sequence MFE | -32.94 |
| Consensus MFE | -22.83 |
| Energy contribution | -22.43 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.536728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 21344557 118 - 23011544 --CAGAACAACAGCACUUUCGAUGGAUGUUUUCAAGUGAGACUUGACAAACAGUAGAGCUACGAUGUGAUAAUUCCAACGGAGGUAUCGAUAGCGCAUGGUCAUCGAUACCUCACGCCAA --..........((.....((.((((((((((((((.....))))).)))))(((....)))...........)))).))(((((((((((..(.....)..)))))))))))..))... ( -36.00, z-score = -2.94, R) >droEre2.scaffold_4845 19436834 111 + 22589142 --CGAAACAACA--ACUUUCGAUGGAUAUUUCCAAGUGGGACUUU----ACAUUAGAGCUGCGAUGUGAUAG-UCCGAUAGGGGUAUCGAUAGCGCAGGGUCAUCGAUGUUUCACGCCAA --.((((((...--....(((((((....((((....))))....----.........(((((...(((((.-(((....))).)))))....)))))..)))))))))))))....... ( -33.51, z-score = -2.03, R) >droYak2.chr2R 19709133 114 + 21139217 --CAGAACAACAACACUUUCGAUGGAUGUUUUCAAGUGGGACAGC----GCAGUAGAGCUGAGAUGUGAUAGUUCCGAUGGGGGUAUCGAUAGCGAAGUGUCAUCGAUGUUUCACGCCAA --..(((((.(.(((((((..((.((((..((((..((((((..(----(((.((....))...))))...)))))).))))..)))).))...)))))))....).)))))........ ( -27.40, z-score = 0.68, R) >droSec1.super_18 1273326 112 - 1342155 CACGGAACAACAACAC--UUGAUGGAUGUUUUCAAGUGGGACUUA------AGUAGAGCUGCGAUGCGAUAGUUCCAAGGGAGGUAUCGAUAGUGCAGAGUCAUCGAUACCUCACGCCAA ...(((((.....(((--((((.........))))))).......------..((..((......))..)))))))..(((((((((((((..(.....)..)))))))))))...)).. ( -36.00, z-score = -2.67, R) >droSim1.chr2L 20945570 114 - 22036055 CUCGGAACAACAACACUUUUGAUGGAUGUUUUCAAGUGGGACUUA------AGUAGAGCUGCGAUGCGAUAGUUCCAAGGGAGGUAUCGAUAGUGCAGAGUCAUCGAUACCUCACGCCAA .((((((.........))))))(((.(((...(...(((((((..------.(((.........)))...)))))))..)(((((((((((..(.....)..))))))))))))))))). ( -31.80, z-score = -1.08, R) >consensus __CGGAACAACAACACUUUCGAUGGAUGUUUUCAAGUGGGACUUA_____CAGUAGAGCUGCGAUGUGAUAGUUCCAAGGGAGGUAUCGAUAGCGCAGAGUCAUCGAUACCUCACGCCAA ......................(((...........(((((((.........(((....)))........)))))))...(((((((((((..(.....)..)))))))))))...))). (-22.83 = -22.43 + -0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:56:16 2011