| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,327,506 – 21,327,601 |
| Length | 95 |
| Max. P | 0.938239 |

| Location | 21,327,506 – 21,327,601 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 71.57 |
| Shannon entropy | 0.59055 |
| G+C content | 0.50702 |
| Mean single sequence MFE | -30.65 |
| Consensus MFE | -12.51 |
| Energy contribution | -13.94 |
| Covariance contribution | 1.43 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.875693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
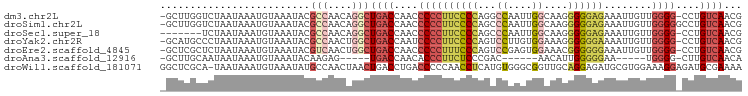
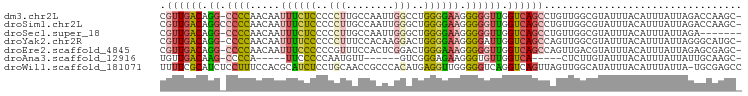
>dm3.chr2L 21327506 95 + 23011544 -GCUUGGUCUAAUAAAUGUAAAUACGCCAACAGGCUGACCAACCCCCUUCCCCAGGCCAAUUGGCAAGGGGGAGAAAUUGUUGGGG-CCUGUCAACG -...((((.((...........)).))))(((((((..(((((...(((((((..(((....)))..))))))).....)))))))-)))))..... ( -36.10, z-score = -2.15, R) >droSim1.chr2L 20928578 96 + 22036055 -GCUUGGUCUAAUAAAUGUAAAUACGCCAACAGGCUGACCAACCCCCUUCCCCAGCCCAAUUGGCAAGGGGGAGAAAUUGUUGGGGGCCUGUCAACG -...((((.((...........)).))))(((((((..(((((...(((((((.(((.....)))..))))))).....))))).)))))))..... ( -34.70, z-score = -1.55, R) >droSec1.super_18 1257548 89 + 1342155 -------UCUAAUAAAUGUAAAUACGCCAACAGGCUGACCAACCCCCUUCCCCAGCCCAAUUGGCAAGGGGGAGAAAUUGUUGGGG-CCUGUCAACG -------.................((...(((((((..(((((...(((((((.(((.....)))..))))))).....)))))))-)))))...)) ( -31.60, z-score = -2.28, R) >droYak2.chr2R 19690662 95 - 21139217 -GCAUGCCCUAAUAAAUGUAAAUACGCCAACUGGCUGACCAAUCCCCUUCCCCAGUCCUUGUGGAAAGGGGGGAAAAUUGUUGGGG-CCUGUCAACG -(((.((((((((((..........(((....)))............((((((..(((....)))...))))))...)))))))))-).)))..... ( -35.20, z-score = -2.65, R) >droEre2.scaffold_4845 19419634 95 - 22589142 -GCUCGCUCUAAUAAAUGUAAAUACGUCAACUGGCUGACCAACCCCCUUUCCCAGUCCGAGUGGAAACGGGGGGAAAUUGUUGGGG-CCUGUCAACG -((..((((((((((..........((((......))))...(((((((((((.......).))))).)))))....)))))))))-)..))..... ( -30.60, z-score = -1.43, R) >droAna3.scaffold_12916 5278048 79 - 16180835 -GCUUGCAAUAAUAAAUGUAAAUACAAGAG-----UGACCAACACCCUUCUCCCGAC------AACAUUGGGGGAA-----UGGGG-CUUGUCAACA -..(((((........))))).........-----((((.((..((((((((((((.------....)))))))))-----.))).-.))))))... ( -22.80, z-score = -2.07, R) >droWil1.scaffold_181071 823290 96 + 1211509 GGCUCGCA-UAAUAAAUGUAAAUAUGCCAACUAACUGACCUGACCCCCAACCUCAUGUGGGCGGUUGCAGGAGAUGCGUGGAAAGGAGAUGCGAAAA ...(((((-(............(((((((......)).(((((((((((........)))).)))..))))....)))))........))))))... ( -23.55, z-score = -0.22, R) >consensus _GCUUGCUCUAAUAAAUGUAAAUACGCCAACAGGCUGACCAACCCCCUUCCCCAGUCCAAGUGGAAAGGGGGGGAAAUUGUUGGGG_CCUGUCAACG .........................(((....)))((((....((((((((((...((....))....))))))........))))....))))... (-12.51 = -13.94 + 1.43)
| Location | 21,327,506 – 21,327,601 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 71.57 |
| Shannon entropy | 0.59055 |
| G+C content | 0.50702 |
| Mean single sequence MFE | -32.46 |
| Consensus MFE | -14.10 |
| Energy contribution | -14.14 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.938239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
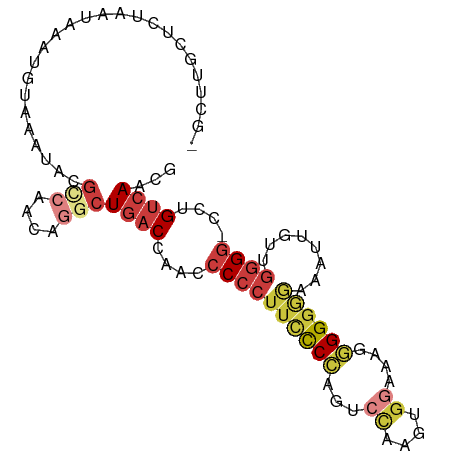
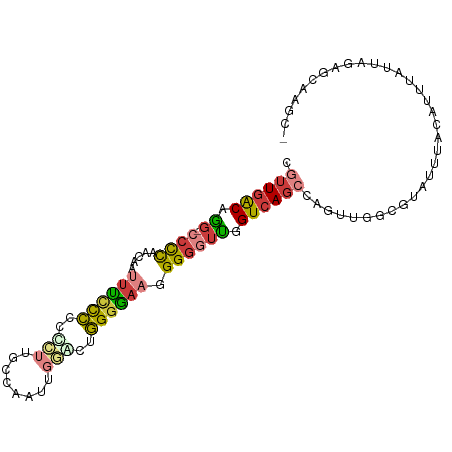
>dm3.chr2L 21327506 95 - 23011544 CGUUGACAGG-CCCCAACAAUUUCUCCCCCUUGCCAAUUGGCCUGGGGAAGGGGGUUGGUCAGCCUGUUGGCGUAUUUACAUUUAUUAGACCAAGC- (((..(((((-(.(((((..(..((.((((..(((....)))..)))).))..))))))...))))))..))).......................- ( -40.80, z-score = -3.21, R) >droSim1.chr2L 20928578 96 - 22036055 CGUUGACAGGCCCCCAACAAUUUCUCCCCCUUGCCAAUUGGGCUGGGGAAGGGGGUUGGUCAGCCUGUUGGCGUAUUUACAUUUAUUAGACCAAGC- (((..((((((..(((((..(..((.((((..(((.....))).)))).))..))))))...))))))..))).......................- ( -40.10, z-score = -2.58, R) >droSec1.super_18 1257548 89 - 1342155 CGUUGACAGG-CCCCAACAAUUUCUCCCCCUUGCCAAUUGGGCUGGGGAAGGGGGUUGGUCAGCCUGUUGGCGUAUUUACAUUUAUUAGA------- (((..(((((-(.(((((..(..((.((((..(((.....))).)))).))..))))))...))))))..))).................------- ( -39.70, z-score = -3.26, R) >droYak2.chr2R 19690662 95 + 21139217 CGUUGACAGG-CCCCAACAAUUUUCCCCCCUUUCCACAAGGACUGGGGAAGGGGAUUGGUCAGCCAGUUGGCGUAUUUACAUUUAUUAGGGCAUGC- ......((.(-(((...(((((..(.((((..(((....)))..))))..)..)))))....(((....)))................)))).)).- ( -32.10, z-score = -1.20, R) >droEre2.scaffold_4845 19419634 95 + 22589142 CGUUGACAGG-CCCCAACAAUUUCCCCCCGUUUCCACUCGGACUGGGAAAGGGGGUUGGUCAGCCAGUUGACGUAUUUACAUUUAUUAGAGCGAGC- .((((((..(-((((.....((((((.(((........)))...)))))).)))))..)))))).......(((.((((.......)))))))...- ( -31.30, z-score = -1.64, R) >droAna3.scaffold_12916 5278048 79 + 16180835 UGUUGACAAG-CCCCA-----UUCCCCCAAUGUU------GUCGGGAGAAGGGUGUUGGUCA-----CUCUUGUAUUUACAUUUAUUAUUGCAAGC- ...((((.((-((((.-----(((.(((......------...))).)))))).))).))))-----..((((((..............)))))).- ( -21.34, z-score = -1.27, R) >droWil1.scaffold_181071 823290 96 - 1211509 UUUUCGCAUCUCCUUUCCACGCAUCUCCUGCAACCGCCCACAUGAGGUUGGGGGUCAGGUCAGUUAGUUGGCAUAUUUACAUUUAUUA-UGCGAGCC ..(((((((.................(((.(((((..........))))).)))....(((((....)))))...............)-)))))).. ( -21.90, z-score = 0.06, R) >consensus CGUUGACAGG_CCCCAACAAUUUCCCCCCCUUGCCAAUUGGACUGGGGAAGGGGGUUGGUCAGCCAGUUGGCGUAUUUACAUUUAUUAGAGCAAGC_ .((((((....((((......((((((.................))))))))))....))))))................................. (-14.10 = -14.14 + 0.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:56:14 2011