
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,310,001 – 21,310,116 |
| Length | 115 |
| Max. P | 0.640399 |

| Location | 21,310,001 – 21,310,116 |
|---|---|
| Length | 115 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 66.27 |
| Shannon entropy | 0.68229 |
| G+C content | 0.49245 |
| Mean single sequence MFE | -33.83 |
| Consensus MFE | -10.06 |
| Energy contribution | -9.59 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.640399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 21310001 115 + 23011544 --UAUUGCGGGUAGCGAGCCCUCACCAUCCCAUCAGGGUUGUCAAGCAUUUGGUGGCUCUGAUAAACAGCACCCUCUUCCUAAAAAUGGCUGAAAGCUGCGAGCCGAAAAACGCAAG --..((((((((.((((....))........((((((((..((((....))))..)))))))).....))))))............(((((..........)))))......)))). ( -34.20, z-score = -0.96, R) >droSim1.chr2L 20910659 115 + 22036055 --UAUUGCGGGUAGCGAGCCCUCGCCAUCAAAUCAGGGUUGUCAAGCAUUUGGCAACUCUGAUAAACAGCACCCUCUUCCUAAAAAUGGCUGUAAGCUGUGAGCCGAAAAGCGCAAU --.(((((((((.((((....))))......((((((((((((((....)))))))))))))).......))))............(((((..........)))))......))))) ( -38.80, z-score = -2.47, R) >droSec1.super_18 1239049 115 + 1342155 --UAUUGCGGGUAGCGAGCCCUCGCCAUCAAAUCAGGGUUGUCAAGCAUUUGGCGACUCUGAUAAACAGCACCAUCUUCCUAAAAAUGGCUGUAAGCUGUGAGCCGAAAAGCGCAAU --.(((((((((.((((....)))).)))..((((((((((((((....))))))))))))))..(((((.....(..((.......))..)...)))))...........)))))) ( -38.10, z-score = -2.10, R) >droEre2.scaffold_4845 19400500 115 - 22589142 --UAUUGCGGGGAGCGAGCCCUCGCCAUCCCAUUAGGGUUGUCAAGCACUCGUCGGUUCUGAUAAACAGCACCCUCUGCCCAAAAAUAGCUGUUAACUGUGAGCCAAAAAACGCAGC --..(((((((((((((....))))..))))....((((.(.....)))))...(((((.....((((((..................))))))......)))))......))))). ( -32.17, z-score = -0.57, R) >droYak2.chr2R 19669597 113 - 21139217 --UAUUGCGGGGAGCGAGCCCUCAC-AGCCCAUCAGGGUUGUCAAGCUUUCGGCGGCUCUGAUAAACA-CACCCUCUGCCUAAAAAUGGCUGUUAACUGUGAGCUGGAAAACGCAAC --..((((((((......)))((.(-(((..((((((((((((........))))))))))))...((-((......(((.......))).......)))).))))))...))))). ( -39.92, z-score = -2.05, R) >droAna3.scaffold_12916 5261861 88 - 16180835 ------GCGGGGAGCGAGCCCUCGCCCACCCAAUAGGGUUGUCAAUAGUUUCCAAAAAUCGAUAACUCGCGCAAGGAUGAGCAAAAAA--CGCAAG--------------------- ------((((((.((((....))))...)))....((((((((.................))))))))((.((....)).))......--)))...--------------------- ( -23.83, z-score = -0.97, R) >dp4.chr4_group4 3447203 114 + 6586962 UAAAAUGCGAGGAGCGAGCCCUCGCCCUCCCACCAGGGCUGCAAGUUUUGGGAUUGCGCUUAUAAACAGCAUAGUAAUGUGCUAAAAAAUACCAAAUAAUGA---CAAAAACGCAAA .....((.((((.((((....)))))))).))(((((((.....)))))))..(((((.........((((((....)))))).........((.....)).---......))))). ( -31.80, z-score = -2.17, R) >droPer1.super_10 2475478 114 + 3432795 UAAAAUGCGAGGAGCGAGCCCUCGCCCUCCCACCAGGGCUGCAAGUUUUGGGAUUGCGCUUAUAAACAGCAUAGUAAUGUGCUAAAAAAUACCAAAUAAUGA---CAAAAACGCAAA .....((.((((.((((....)))))))).))(((((((.....)))))))..(((((.........((((((....)))))).........((.....)).---......))))). ( -31.80, z-score = -2.17, R) >consensus __UAUUGCGGGGAGCGAGCCCUCGCCAUCCCAUCAGGGUUGUCAAGCAUUUGGCGGCUCUGAUAAACAGCACCCUCUUCCUAAAAAUAGCUGUAAACUGUGAGCCGAAAAACGCAAA .....((((....((.((((((............))))))))..............................................((........))...........)))).. (-10.06 = -9.59 + -0.47)
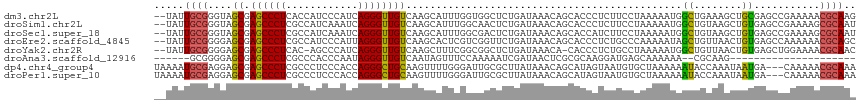

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:56:12 2011