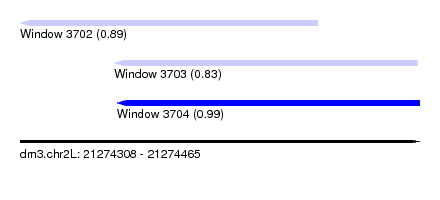
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,274,308 – 21,274,465 |
| Length | 157 |
| Max. P | 0.991656 |

| Location | 21,274,308 – 21,274,425 |
|---|---|
| Length | 117 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.66 |
| Shannon entropy | 0.57328 |
| G+C content | 0.63387 |
| Mean single sequence MFE | -51.03 |
| Consensus MFE | -17.88 |
| Energy contribution | -17.98 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.71 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.893091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 21274308 117 - 23011544 CUCCUUUGUGGUGGAAACGGGUCGGGUUCCGGGUAACGACUGAUUACCGGGGAUGCCGCCCAAAGUGACUCCACUACCGCCCAAGGAAACGGAUGCGCCACCGCUUCCGCCACCUAU--- .(((((.(.(((((....(((.((((((((.(((((.......))))).))))).))))))..((((....)))).)))))))))))..((((.(((....))).))))........--- ( -48.40, z-score = -2.09, R) >droEre2.scaffold_4845 19366452 117 + 22589142 CUCCUUUGUGGUGGAAACGGGUCGCGUUCCGGGUAGCGACUGAUUGCCGGGGAUGCCGCCCAAAGUGGCUCCACUACCGCCCAAGGGAACGGAUGCACCACCGCUUCCACUACCUAU--- .(((((((.(((((....((((.(((((((.(((((.......))))).))))))).))))..((((....)))).))))))))))))..(((.((......)).))).........--- ( -54.40, z-score = -3.38, R) >droYak2.chr2R 19639360 117 + 21139217 CUCCUUUGUGGUGGAAACGGGUCGCGUUCCGGGUAGCGACUGAUUGCCGGGGAUGCCGCCCAAAGUGGCUCCACUACCACCCAAUGGAGCAGAUGCCACACCGCUUCCACUACCUAU--- .......(((((((((.(((((.(((((((.(((((.......))))).))))))).))))...(((((((..((.(((.....)))))..)).)))))...).)))))))))....--- ( -51.80, z-score = -3.42, R) >droSec1.super_18 1203327 117 - 1342155 CUCCUUUGUGGUGGAAACGGGUCGCGUUCCGGGUAACGACUGAUUACCGGGGAUGCCGCCCAAAGUGACUCCACUACCGCCCAAGGAAACGGAUGCGCCACCGCUUCCGCUUCCUAU--- ......((.(((((....((((.(((((((.(((((.......))))).))))))).))))..((((....)))).)))))))(((((.((((.(((....))).)))).)))))..--- ( -54.00, z-score = -4.34, R) >droSim1.chr2L 20870638 117 - 22036055 CUCCUUUGUGGUGGAAACGGGUCGCGUUCCGGGUAACGACUGAUUACCGGGGAUGCCGCCCAAAGUGACUCCACUACCGCCCAAGGAAACGGAUGCGCCACCGCUUCCGCUUCCUAU--- ......((.(((((....((((.(((((((.(((((.......))))).))))))).))))..((((....)))).)))))))(((((.((((.(((....))).)))).)))))..--- ( -54.00, z-score = -4.34, R) >droAna3.scaffold_12916 5236283 117 + 16180835 CUCCUUUGUGUUUGAAACUGUCCGACCUGCCUGCAUCCCUUGGGUGCUGGCCACCCCGCCCAAAGUGGCGCCACUGCCUCCCAAUGGAAUCGCCGAUCCGCCACUGCCAGCGCCAAU--- .......(((((.(....((..((....(((.((((((...)))))).))).....))..)).(((((((...(.((.(((....)))...)).)...))))))).).)))))....--- ( -32.20, z-score = -0.31, R) >dp4.chr4_group4 3411675 117 - 6586962 CUCCUUUGUGGAUGAAACGGGUCGCGUUCCCGGCAGCGACUGGUUGCUGGAGACGCCUCCCAGUGUGGCACCACUGCCGCCCAGGGGAAUGGCAUUGCCUCCGCUCCCACUUCCGAU--- .......((((.......(((..((((..((((((((.....))))))))..))))..)))((((.((((....((((.((....))...)))).))))..)))).)))).......--- ( -50.80, z-score = -1.82, R) >droPer1.super_10 2439902 117 - 3432795 CUCCUUUGUGGAUGAAACGGGUCGCGUUCCCGGCAGCGACUGGUUGCUGGAGACGCCUCCCAGUGUGGCGCCACUGCCGCCCAGGGGAAUGGCAUUGCCUCCGCUACCACUGCCGAU--- .......((((.......(((..((((..((((((((.....))))))))..))))..)))...((((.(.((.((((.((....))...)))).)).).))))..)))).......--- ( -51.10, z-score = -1.49, R) >droWil1.scaffold_181071 757991 117 - 1211509 UUCCCUUACGGCAGAUACUGGUCUUGUGCCAGGCACCGAUUGUAAACUGGAUACGCCACCCAAUGUGGCUCCACUUCCGCCCAGGGGGAUGGCUGUACCUCCACUGCCGCUACCUAU--- ........((((((...(((((.....)))))...............((((((((((((((..((.(((.........)))))..))).)))).)))..))))))))))........--- ( -40.90, z-score = -1.46, R) >droMoj3.scaffold_6500 21151568 117 + 32352404 AUCCCUGGCGGUGGAGACGGGUCGCGUGCCCGCUGCACUCACUGUGCUGGCCACGCCGCCCAGGGUGGCGCCACUGCCACCCAGAGGUAUGGCUGUGCCGCCACUGCCGCUGCCCAU--- ......((((((((....((((.(((((.(((..((((.....))))))).))))).))))..(((((((.(((.((((((....)))..))).))).))))))).))))))))...--- ( -64.00, z-score = -2.18, R) >droVir3.scaffold_12723 187573 117 + 5802038 CUCCCUUGUGGCGGAGACGGGGCGCGUGACCGACGCCGACAUUGUGCUGGCCACACCGCCCAAAGUGGCGCCACUGCCGCCCAGAGGAAUGGCUGUGCCGCCACUGCCGCUGCCCAU--- .......(..((((.....(((((.(((.(((.(((.......))).))).)))..)))))..(((((((.(((.(((.((....))...))).))).))))))).))))..)....--- ( -52.50, z-score = -1.28, R) >droGri2.scaffold_15126 7561571 120 - 8399593 CUCCUUUGUGCUGGAGGCGGGGCGUGUGCCCGGCGCAGUCAUUGGGAUGGCAACGCCGCCCAAAGUGGCGCCACUGCCGCCCAGGGGAAUGGCGGAGCCACCACUGCUGGCACCACCAAU .(((((((....((.((((((((....))))(((((...(.(((((.((((...))))))))).)..)))))..)))).))))))))).(((.((.((((.......)))).)).))).. ( -58.30, z-score = -0.53, R) >consensus CUCCUUUGUGGUGGAAACGGGUCGCGUUCCCGGCACCGACUGAUUGCUGGGGACGCCGCCCAAAGUGGCUCCACUGCCGCCCAAGGGAAUGGCUGCGCCACCGCUGCCGCUACCUAU___ .......(((((......(((..(((((((.(.(((.......)))).))).))))..)))...(((....))).))))).........((((.((......)).))))........... (-17.88 = -17.98 + 0.10)
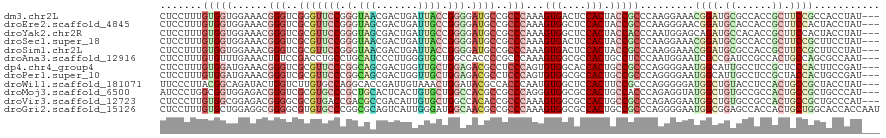
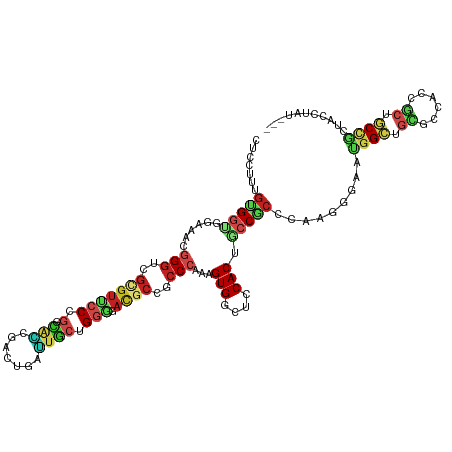
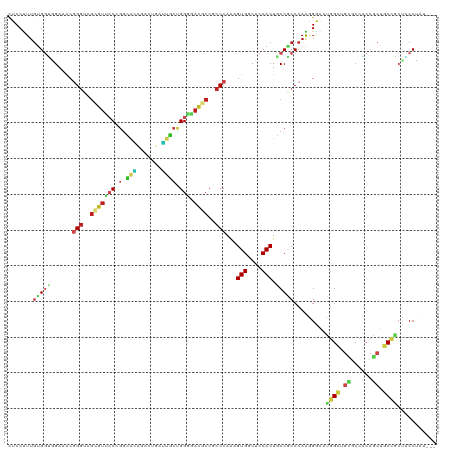
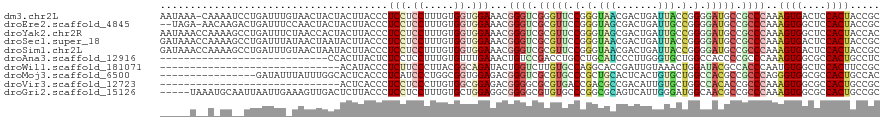
| Location | 21,274,345 – 21,274,464 |
|---|---|
| Length | 119 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.22 |
| Shannon entropy | 0.72377 |
| G+C content | 0.57256 |
| Mean single sequence MFE | -34.80 |
| Consensus MFE | -15.17 |
| Energy contribution | -15.32 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.65 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.834389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 21274345 119 - 23011544 AAUAAA-CAAAAUCCUGAUUUGUAACUACUACUUACCCUCCUCCUUUGUGGUGGAAACGGGUCGGGUUCCGGGUAACGACUGAUUACCGGGGAUGCCGCCCAAAGUGACUCCACUACCGC .....(-((((.......)))))........................((((((((.(((((.((((((((.(((((.......))))).))))).))))))...))...))))))))... ( -33.50, z-score = -0.52, R) >droEre2.scaffold_4845 19366489 117 + 22589142 --UAGA-AACAAGACUGAUUUCCAACUACUACUUACCCUCCUCCUUUGUGGUGGAAACGGGUCGCGUUCCGGGUAGCGACUGAUUGCCGGGGAUGCCGCCCAAAGUGGCUCCACUACCGC --....-...........((((((.((((..................)))))))))).((((.(((((((.(((((.......))))).))))))).))))..((((....))))..... ( -37.97, z-score = -0.97, R) >droYak2.chr2R 19639397 120 + 21139217 AAUAAACCAAAAGCCUGAUUUCUAACCACUACUUACCCUCCUCCUUUGUGGUGGAAACGGGUCGCGUUCCGGGUAGCGACUGAUUGCCGGGGAUGCCGCCCAAAGUGGCUCCACUACCAC ...........((((..........(((((((...............)))))))....((((.(((((((.(((((.......))))).))))))).)))).....)))).......... ( -40.16, z-score = -1.93, R) >droSec1.super_18 1203364 120 - 1342155 GAUAAACCAAAAGCCUGAUUUAUAACUAAUACUUACCCUCCUCCUUUGUGGUGGAAACGGGUCGCGUUCCGGGUAACGACUGAUUACCGGGGAUGCCGCCCAAAGUGACUCCACUACCGC .(((((.((......)).)))))........................((((((((.((((((.(((((((.(((((.......))))).))))))).))))...))...))))))))... ( -36.60, z-score = -2.07, R) >droSim1.chr2L 20870675 120 - 22036055 GAUAAACCAAAAGCCUGAUUUGUAACUAAUACUUACCCUCCUCCUUUGUGGUGGAAACGGGUCGCGUUCCGGGUAACGACUGAUUACCGGGGAUGCCGCCCAAAGUGACUCCACUACCGC .(((((.((......)).)))))........................((((((((.((((((.(((((((.(((((.......))))).))))))).))))...))...))))))))... ( -36.60, z-score = -1.71, R) >droAna3.scaffold_12916 5236320 92 + 16180835 ----------------------------CCACUUACUCUCCUCCUUUGUGUUUGAAACUGUCCGACCUGCCUGCAUCCCUUGGGUGCUGGCCACCCCGCCCAAAGUGGCGCCACUGCCUC ----------------------------...............(((((.(((.((.....)).)))..(((.((((((...)))))).))).........))))).((((....)))).. ( -20.50, z-score = -0.28, R) >droWil1.scaffold_181071 758028 90 - 1211509 ------------------------------ACAUACCCUCUUCCCUUACGGCAGAUACUGGUCUUGUGCCAGGCACCGAUUGUAAACUGGAUACGCCACCCAAUGUGGCUCCACUUCCGC ------------------------------..................(((.((...(((((.....)))))...............((((...(((((.....))))))))))).))). ( -21.20, z-score = -0.37, R) >droMoj3.scaffold_6500 21151605 104 + 32352404 ----------------GAUAUUUAUUUGGCACUCACCCUCAUCCCUGGCGGUGGAGACGGGUCGCGUGCCCGCUGCACUCACUGUGCUGGCCACGCCGCCCAGGGUGGCGCCACUGCCAC ----------------..........((((.(.((((((..((((.....).)))...((((.(((((.(((..((((.....))))))).))))).))))))))))).))))....... ( -44.40, z-score = -1.17, R) >droVir3.scaffold_12723 187610 90 + 5802038 ------------------------------ACUCACCCUCCUCCCUUGUGGCGGAGACGGGGCGCGUGACCGACGCCGACAUUGUGCUGGCCACACCGCCCAAAGUGGCGCCACUGCCGC ------------------------------.......((((.((.....)).))))...(((((.(((.(((.(((.......))).))).)))..)))))...((((((....)))))) ( -32.70, z-score = 0.30, R) >droGri2.scaffold_15126 7561611 115 - 8399593 -----UAAAUGCAAUUAAUUGAAAGUUGACUCUUACCCUCCUCCUUUGUGCUGGAGGCGGGGCGUGUGCCCGGCGCAGUCAUUGGGAUGGCAACGCCGCCCAAAGUGGCGCCACUGCCGC -----.....((..(((((.....)))))......(((.(((((........))))).)))))(((.((..(((((...(.(((((.((((...))))))))).)..)))))...))))) ( -44.40, z-score = -1.02, R) >consensus _____A__A_AA__CUGAUUU_UAACUACUACUUACCCUCCUCCUUUGUGGUGGAAACGGGUCGCGUUCCGGGUACCGACUGAUUGCCGGGGACGCCGCCCAAAGUGGCUCCACUACCGC ......................................(((.((.....)).)))...((((.(((((.(.(.(((.......))).).).))))).))))..((((....))))..... (-15.17 = -15.32 + 0.15)
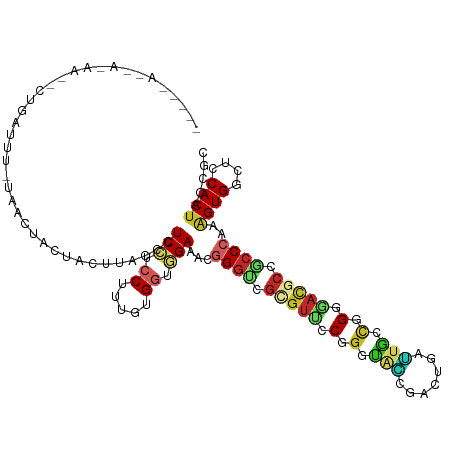
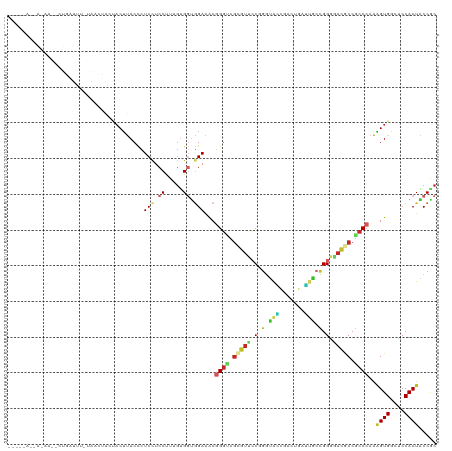
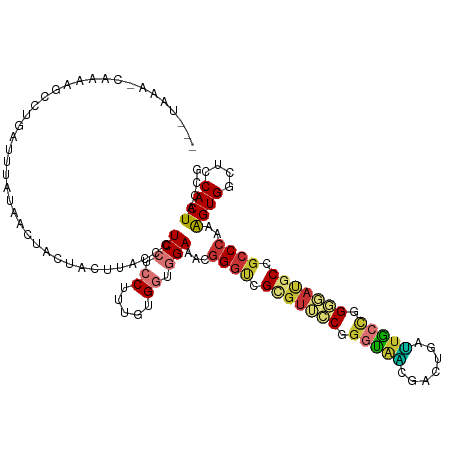
| Location | 21,274,346 – 21,274,465 |
|---|---|
| Length | 119 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.07 |
| Shannon entropy | 0.64031 |
| G+C content | 0.55875 |
| Mean single sequence MFE | -36.65 |
| Consensus MFE | -26.06 |
| Energy contribution | -26.42 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.991656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 21274346 119 - 23011544 CAAUAAA-CAAAAUCCUGAUUUGUAACUACUACUUACCCUCCUCCUUUGUGGUGGAAACGGGUCGGGUUCCGGGUAACGACUGAUUACCGGGGAUGCCGCCCAAAGUGACUCCACUACCG ......(-((((.......)))))........................((((((((.(((((.((((((((.(((((.......))))).))))).))))))...))...)))))))).. ( -33.50, z-score = -0.68, R) >droEre2.scaffold_4845 19366490 117 + 22589142 ---CUAGAAACAAGACUGAUUUCCAACUACUACUUACCCUCCUCCUUUGUGGUGGAAACGGGUCGCGUUCCGGGUAGCGACUGAUUGCCGGGGAUGCCGCCCAAAGUGGCUCCACUACCG ---................((((((.((((..................)))))))))).((((.(((((((.(((((.......))))).))))))).))))..((((....)))).... ( -37.97, z-score = -1.22, R) >droYak2.chr2R 19639398 120 + 21139217 CAAUAAACCAAAAGCCUGAUUUCUAACCACUACUUACCCUCCUCCUUUGUGGUGGAAACGGGUCGCGUUCCGGGUAGCGACUGAUUGCCGGGGAUGCCGCCCAAAGUGGCUCCACUACCA ............((((..........(((((((...............)))))))....((((.(((((((.(((((.......))))).))))))).)))).....))))......... ( -40.16, z-score = -1.82, R) >droSec1.super_18 1203365 120 - 1342155 CGAUAAACCAAAAGCCUGAUUUAUAACUAAUACUUACCCUCCUCCUUUGUGGUGGAAACGGGUCGCGUUCCGGGUAACGACUGAUUACCGGGGAUGCCGCCCAAAGUGACUCCACUACCG ..(((((.((......)).)))))........................((((((((.((((((.(((((((.(((((.......))))).))))))).))))...))...)))))))).. ( -36.60, z-score = -2.13, R) >droSim1.chr2L 20870676 120 - 22036055 CGAUAAACCAAAAGCCUGAUUUGUAACUAAUACUUACCCUCCUCCUUUGUGGUGGAAACGGGUCGCGUUCCGGGUAACGACUGAUUACCGGGGAUGCCGCCCAAAGUGACUCCACUACCG ..(((((.((......)).)))))........................((((((((.((((((.(((((((.(((((.......))))).))))))).))))...))...)))))))).. ( -36.60, z-score = -1.77, R) >droAna3.scaffold_12916 5236321 91 + 16180835 -----------------------------CCACUUACUCUCCUCCUUUGUGUUUGAAACUGUCCGACCUGCCUGCAUCCCUUGGGUGCUGGCCACCCCGCCCAAAGUGGCGCCACUGCCU -----------------------------...............(((((.(((.((.....)).)))..(((.((((((...)))))).))).........))))).((((....)))). ( -20.50, z-score = -0.22, R) >droMoj3.scaffold_6500 21151606 103 + 32352404 -----------------GAUAUUUAUUUGGCACUCACCCUCAUCCCUGGCGGUGGAGACGGGUCGCGUGCCCGCUGCACUCACUGUGCUGGCCACGCCGCCCAGGGUGGCGCCACUGCCA -----------------..........((((.(.((((((..((((.....).)))...((((.(((((.(((..((((.....))))))).))))).))))))))))).))))...... ( -44.40, z-score = -1.08, R) >droGri2.scaffold_15126 7561612 114 - 8399593 ------UAAAUGCAAUUAAUUGAAAGUUGACUCUUACCCUCCUCCUUUGUGCUGGAGGCGGGGCGUGUGCCCGGCGCAGUCAUUGGGAUGGCAACGCCGCCCAAAGUGGCGCCACUGCCG ------...((((..(((((.....)))))......(((.(((((........))))).)))))))(.((..(((((...(.(((((.((((...))))))))).)..)))))...))). ( -43.50, z-score = -1.14, R) >consensus ___UAAA_CAAAAGCCUGAUUUAUAACUACUACUUACCCUCCUCCUUUGUGGUGGAAACGGGUCGCGUUCCGGGUAACGACUGAUUGCCGGGGAUGCCGCCCAAAGUGGCUCCACUACCG .......................................(((.((.....)).)))...((((.(((((((.(((((.......))))).))))))).))))..((((....)))).... (-26.06 = -26.42 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:56:10 2011